A cross-population atlas of genetic associations for 220 human phenotypes
Published in Nature Genetics, 2021
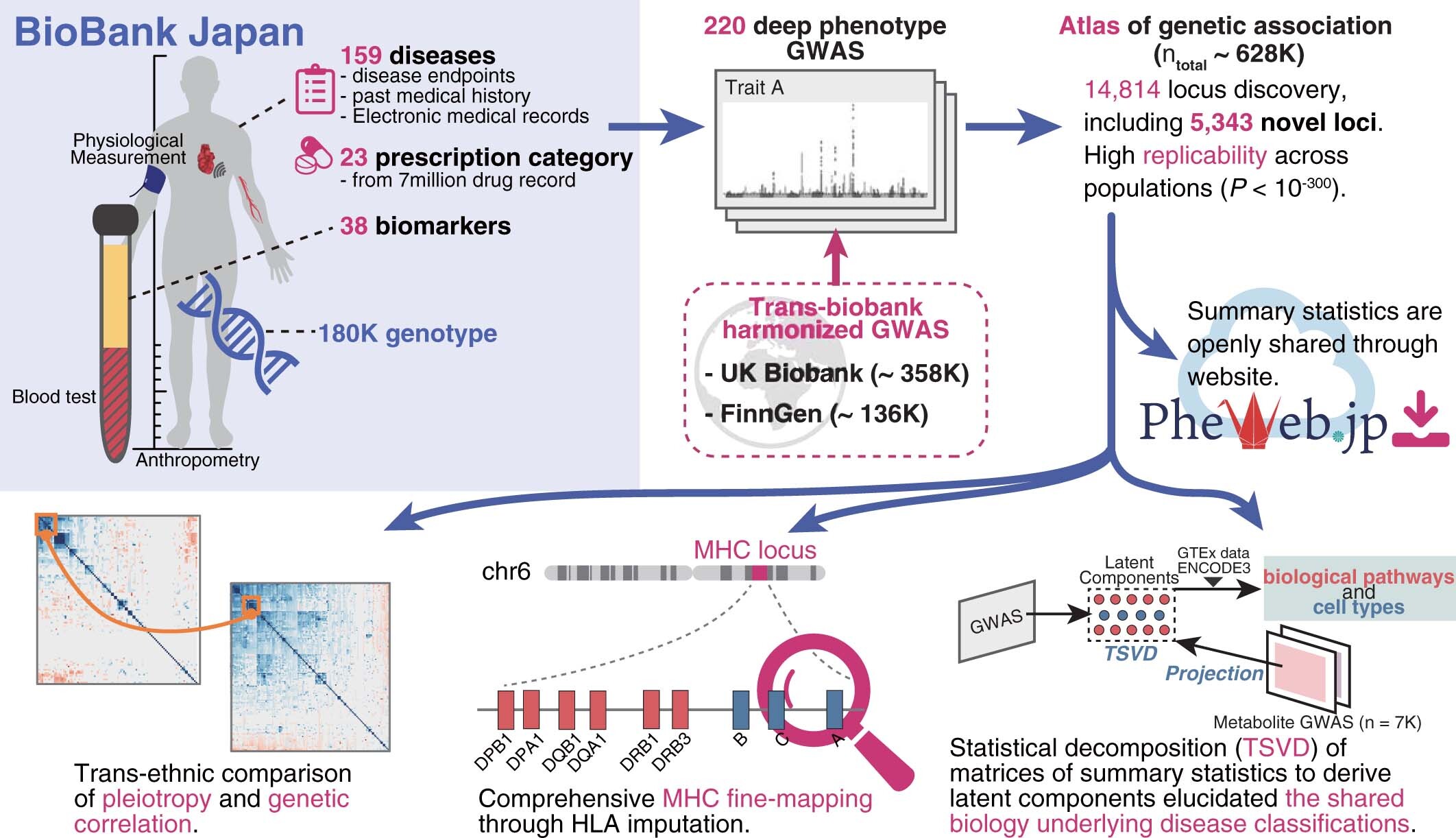
We defined 220 disease endpoints in Biobank Japan by mining the datasets in electronic health records and performed GWAS meta-analysis with the corresponding phenotypes in European populations, UK Biobank, and FinnGen.
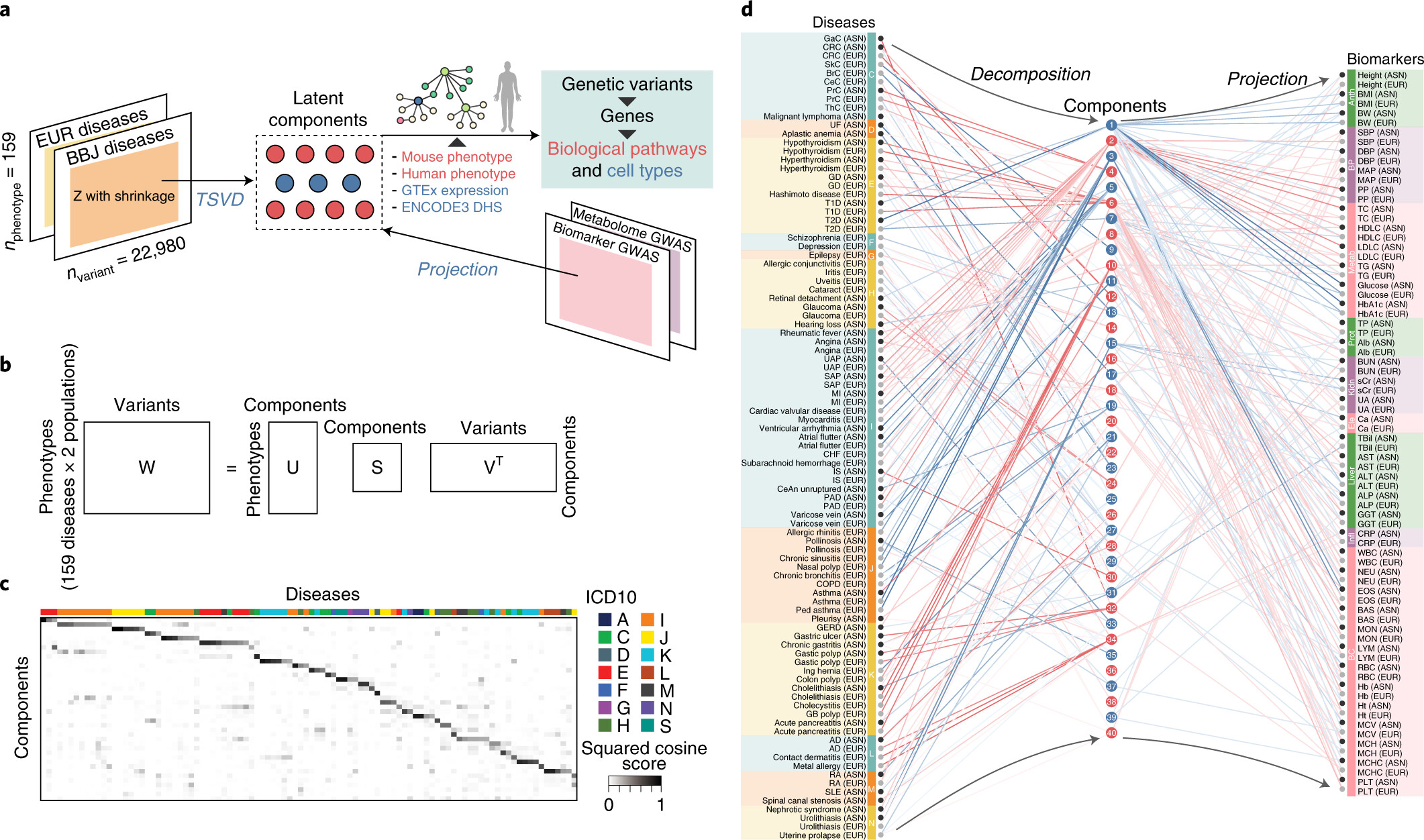
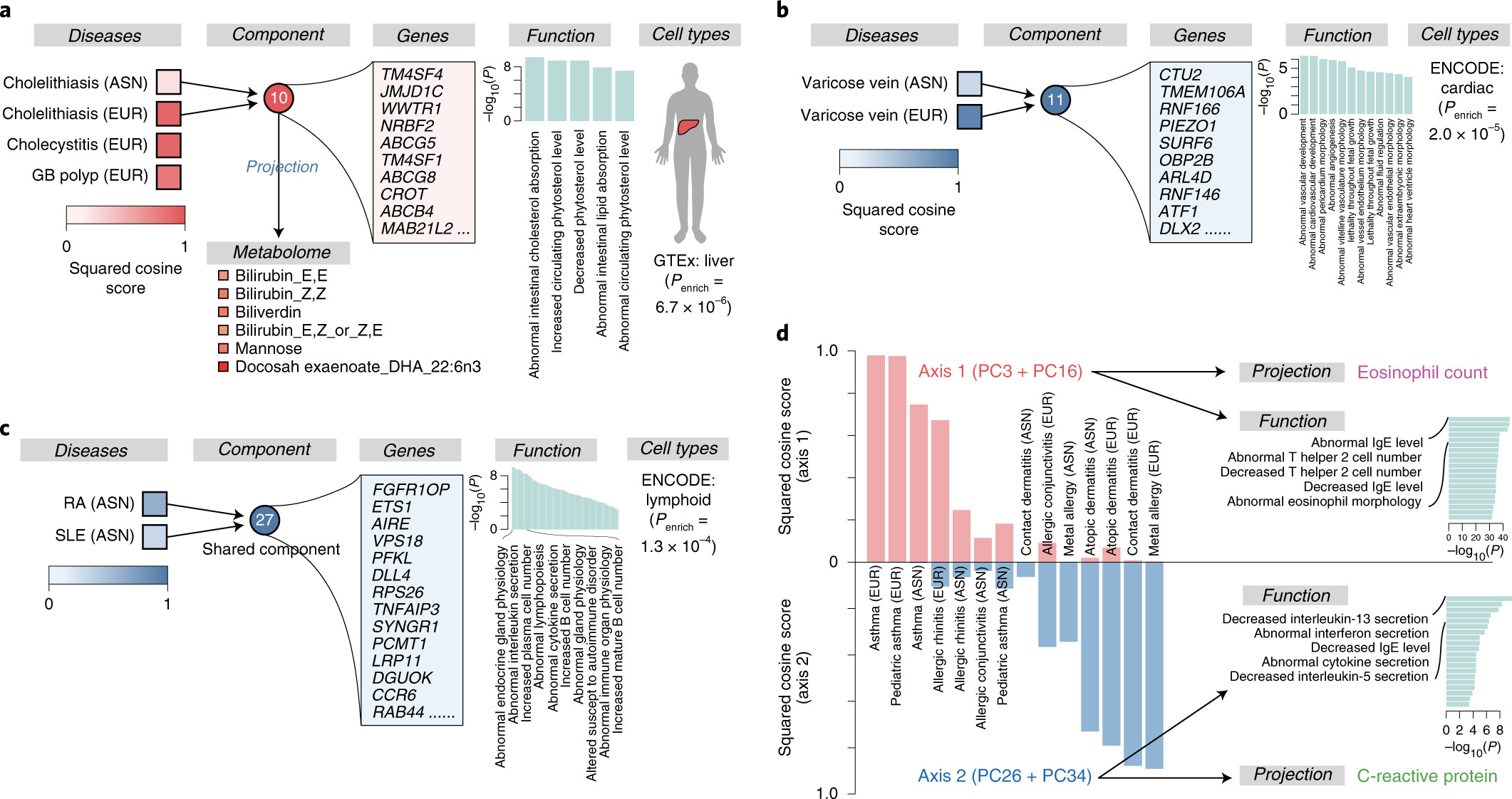
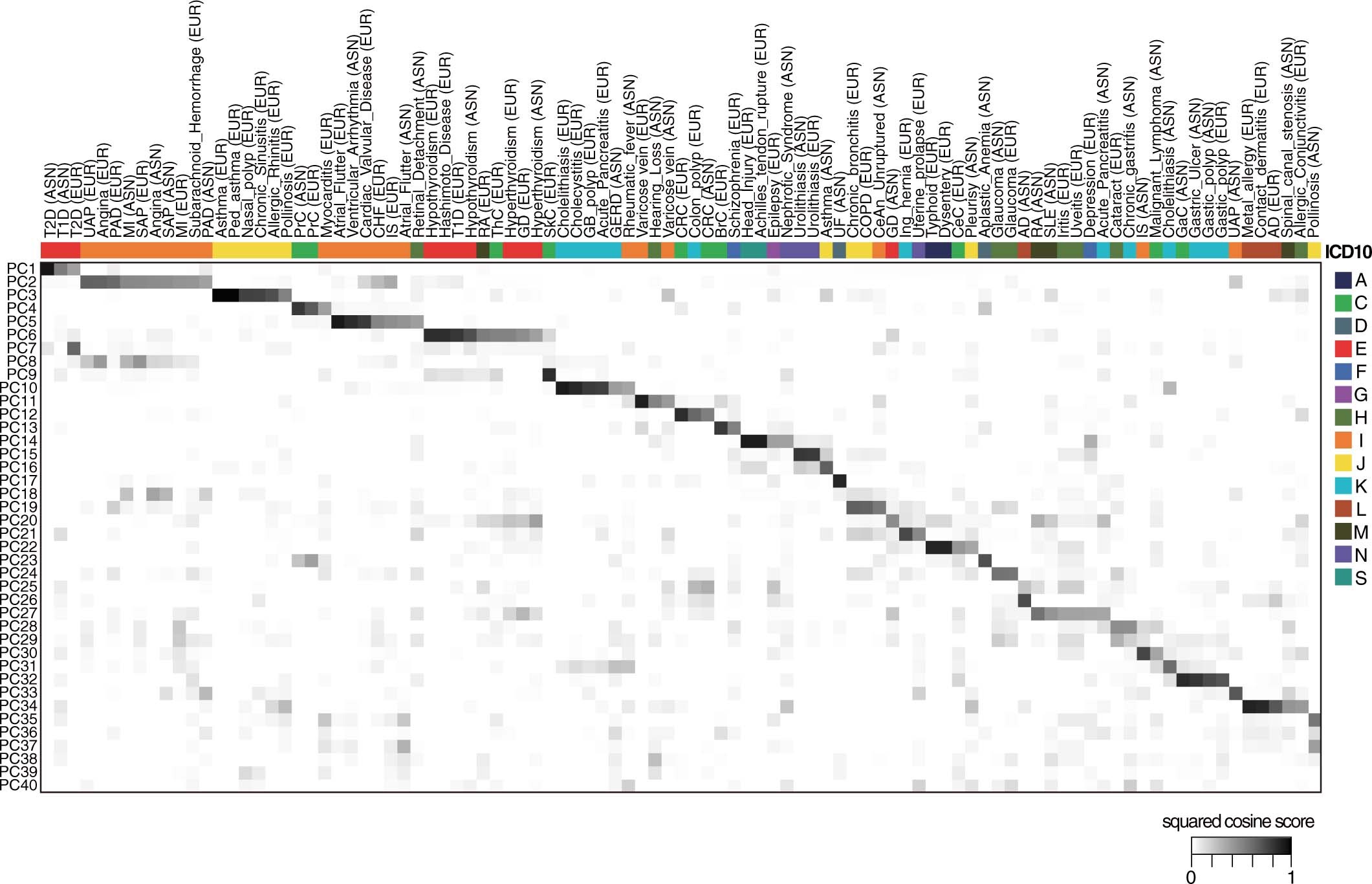
Using a set of GWAS summary statistics of diseases characterized from both European (UK Biobank and FinnGen) and East Asian (Biobank Japan) populations, we dissected latent DeGAs components of multi-ethnic association summary statistics. We annotated each component by pathway and cell-type enrichment as well as projection of metabolomic and biomarker summary statistics. We demonstrate how we can use such trans-ethnic annotated latent components to classify diseases based on their genetic basis.
This is a project co-led by Saori and Masa. Yosuke contributed to the design and interpretation of the decomposition analysis.
This paper was highlighted as the cover of the journal. Huge congratulations to Saori, Masa, and the team!

Reference: S. Sakaue*, M. Kanai*, Y. Tanigawa, J. Karjalainen, M. Kurki, S. Koshiba, A. Narita, T. Konuma, K. Yamamoto, M. Akiyama, K. Ishigaki, A. Suzuki, K. Suzuki, W. Obara, K. Yamaji, K. Takahashi, S. Asai, Y. Takahashi, T. Suzuki, N. Sinozaki, H. Yamaguchi, S. Minami, S. Murayama, K. Yoshimori, S. Nagayama, D. Obata, M. Higashiyama, A. Masumoto, Y. Koretsune, F. Gen, K. Ito, C. Terao, T. Yamauchi, I. Komuro, T. Kadowaki, G. Tamiya, M. Yamamoto, Y. Nakamura, M. Kubo, Y. Murakami, K. Yamamoto, Y. Kamatani, A. Palotie, M. A. Rivas, M. Daly, K. Matsuda, Y. Okada, A cross-population atlas of genetic associations for 220 human phenotypes. Nat Gen. 53(10), 1415-1424 (2021). https://doi.org/10.1038/s41588-021-00931-x (full text)
