[Preprint] PRISM: ancestry-aware integration of tissue-specific genomic annotations enhances the transferability of polygenic scores.
Preprint posted on bioRxiv, 2025
In this study, led by Xiaohe (Lucy) Tian, we showed that ancestry-aware integration of tissue-specific genomic annotations enhances the transferability of polygenic scores (PGS).
Abstract
The limited transferability of polygenic scores (PGS) across populations constrains their clinical utility and risks exacerbating health disparities, given challenges in multi-ancestry training, fine-mapping, and variant prioritization using genomic annotations, particularly when biologically relevant reference resources are sparse or unavailable for the target population. Here, we introduce PRISM, a transfer learning approach that jointly addresses these challenges to enhance PGS transferability. Applying PRISM to 7352 fine-mapped variants, 414 ENCODE annotations, and 406,659 individuals from the UK Biobank, we demonstrate that ancestry-aware integration of tissue-specific annotations yields the largest gains in predictive performance for African ancestry, with an average improvement of 13.10% (p=1.6×10−5) over annotation-agnostic multi-ancestry PGS. Notably, the best-performing model uses 102-fold fewer annotations than non-specific models, with contributions from broad categories of annotations. Overall, PRISM complements ongoing data diversification efforts by providing an immediately applicable strategy based on the integration of biologically aligned, best-available resources to address genomic health equity.

Highlights
Accurate genetic prediction of complex traits has the potential to substantially reduce the disease burden, but the limited transferability of polygenic score (PGS) across genetic ancestry groups remains a major challenge.
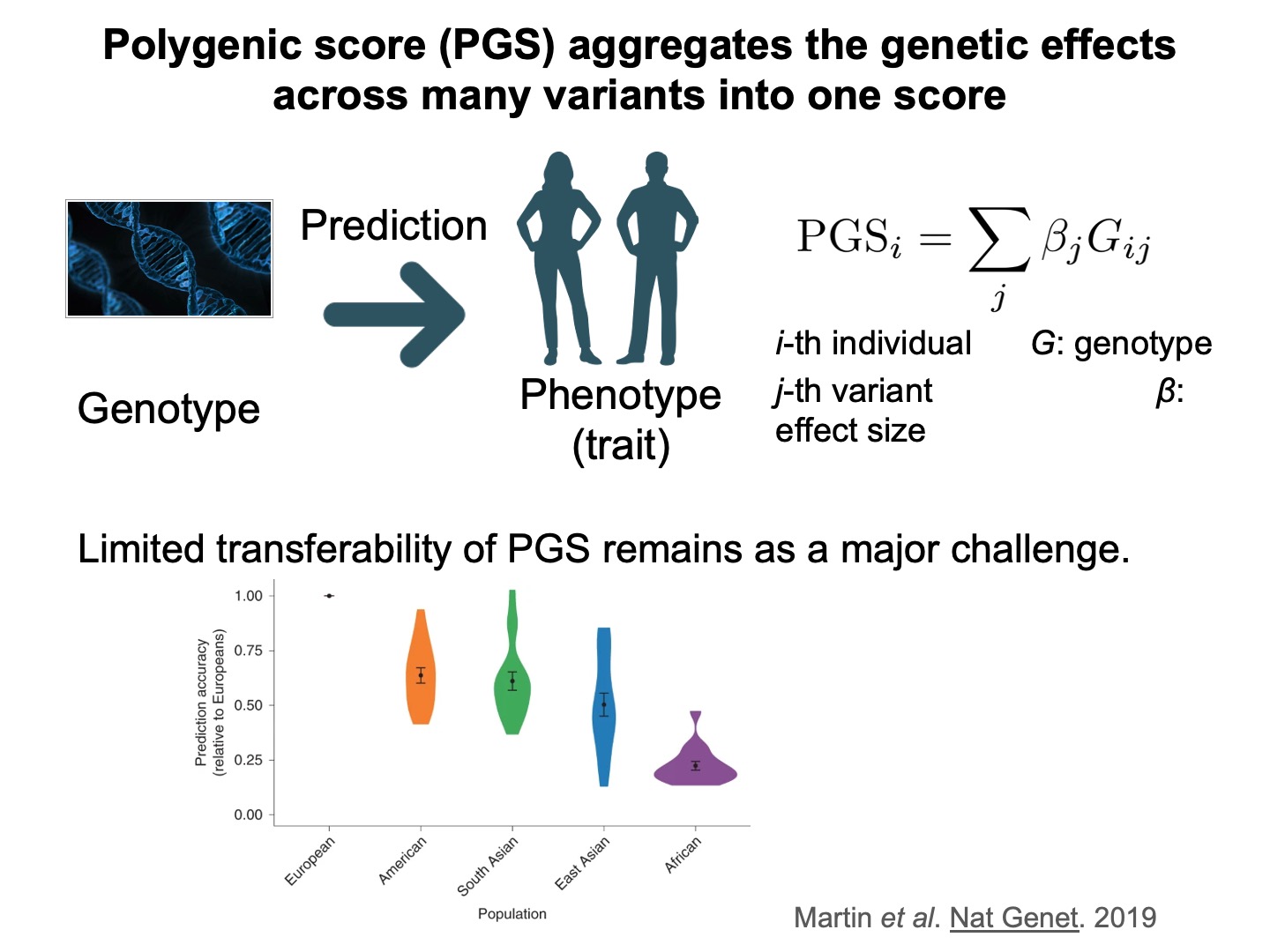
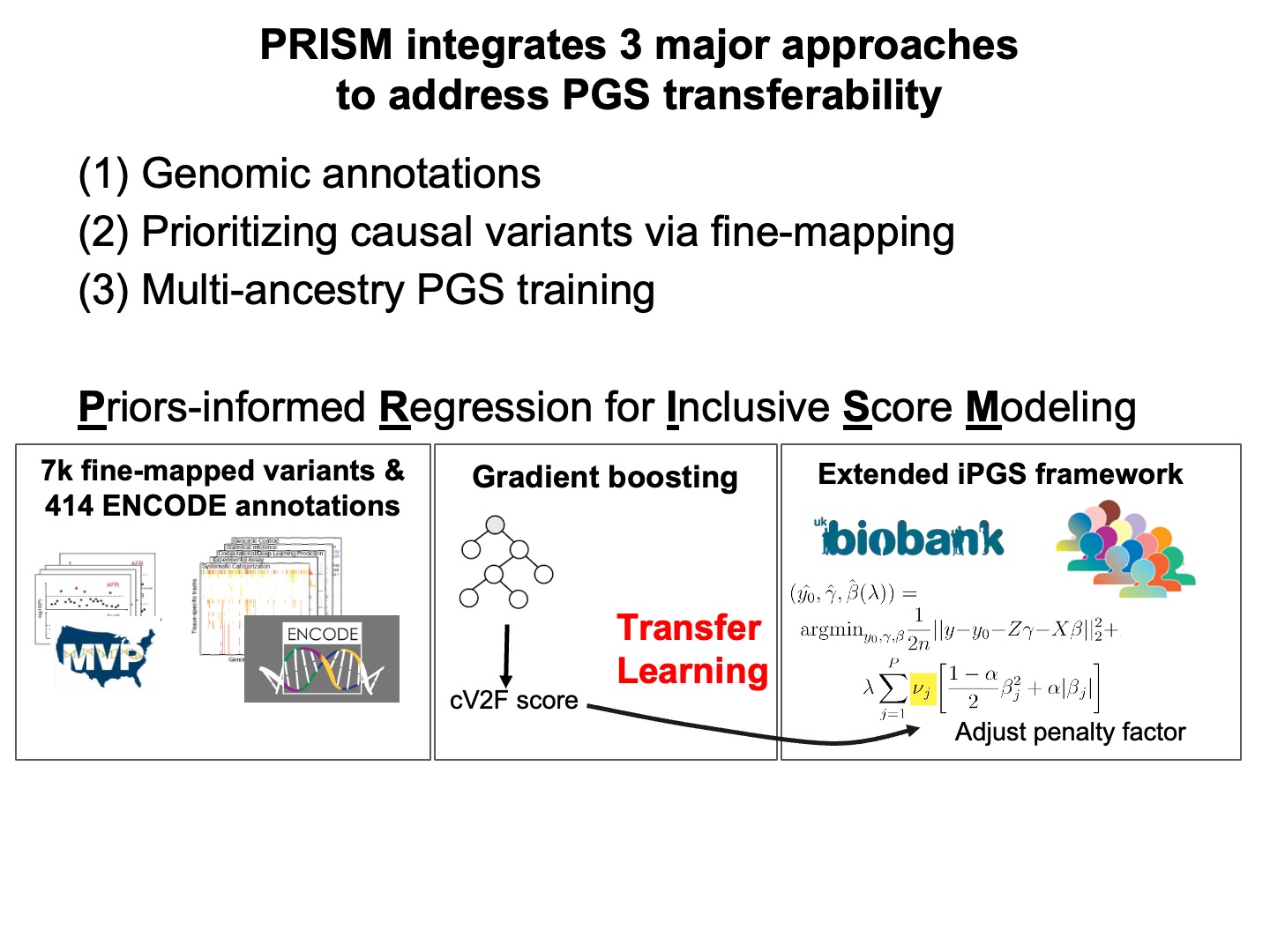
Here we propose to integrate three major approaches to address the PGS transferability. We develop PRISM and apply it to 7352 fine-mapped variants from MVP, 414 ENCODE annotations, and 406,659 individuals from the UK Biobank.
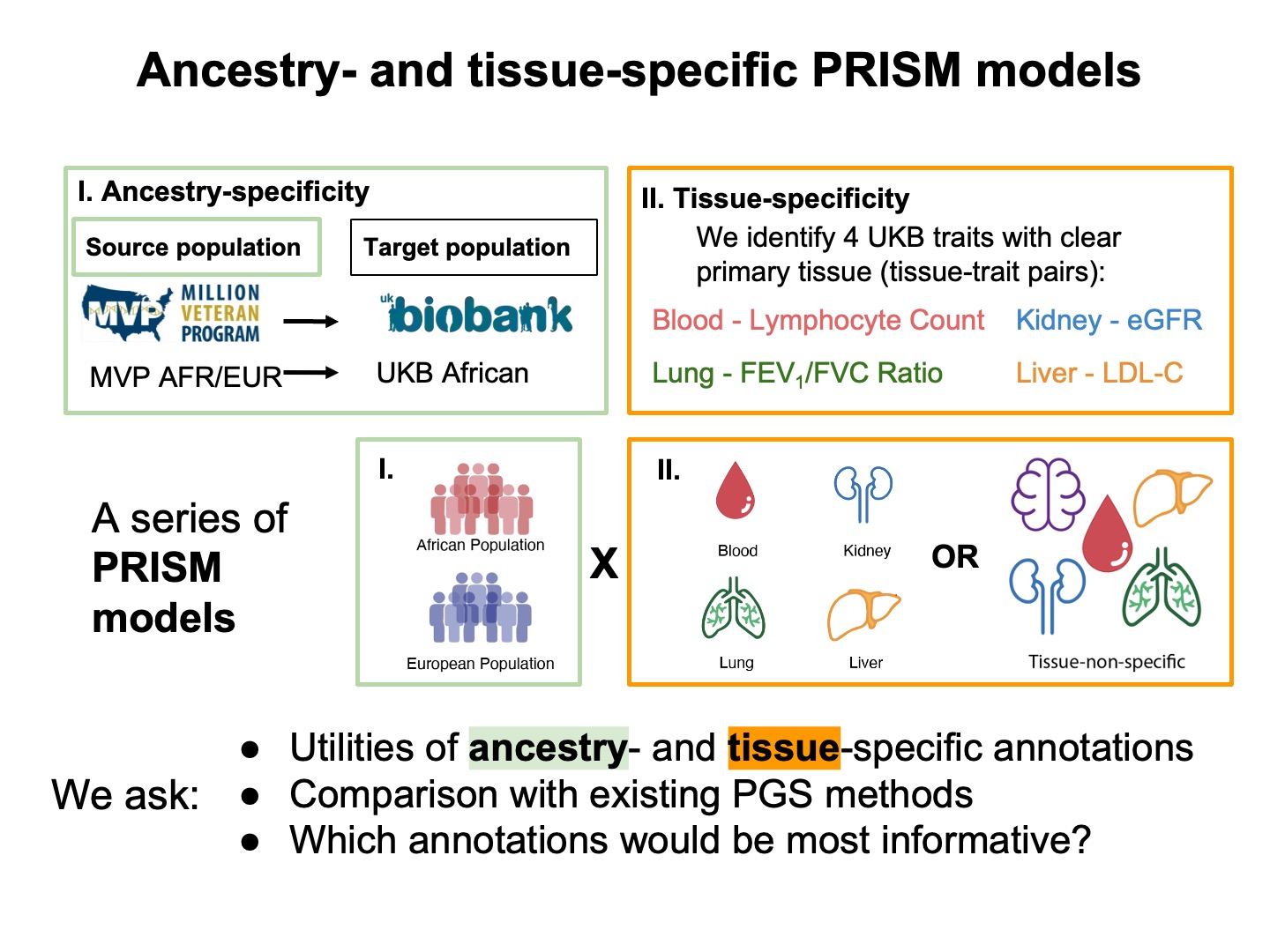
Using PRISM, we evaluated the effects of ancestry and tissue-specific integration of genomic annotations. We also compared our approach with existing methods and investigated which genomic annotation would be the most informative.
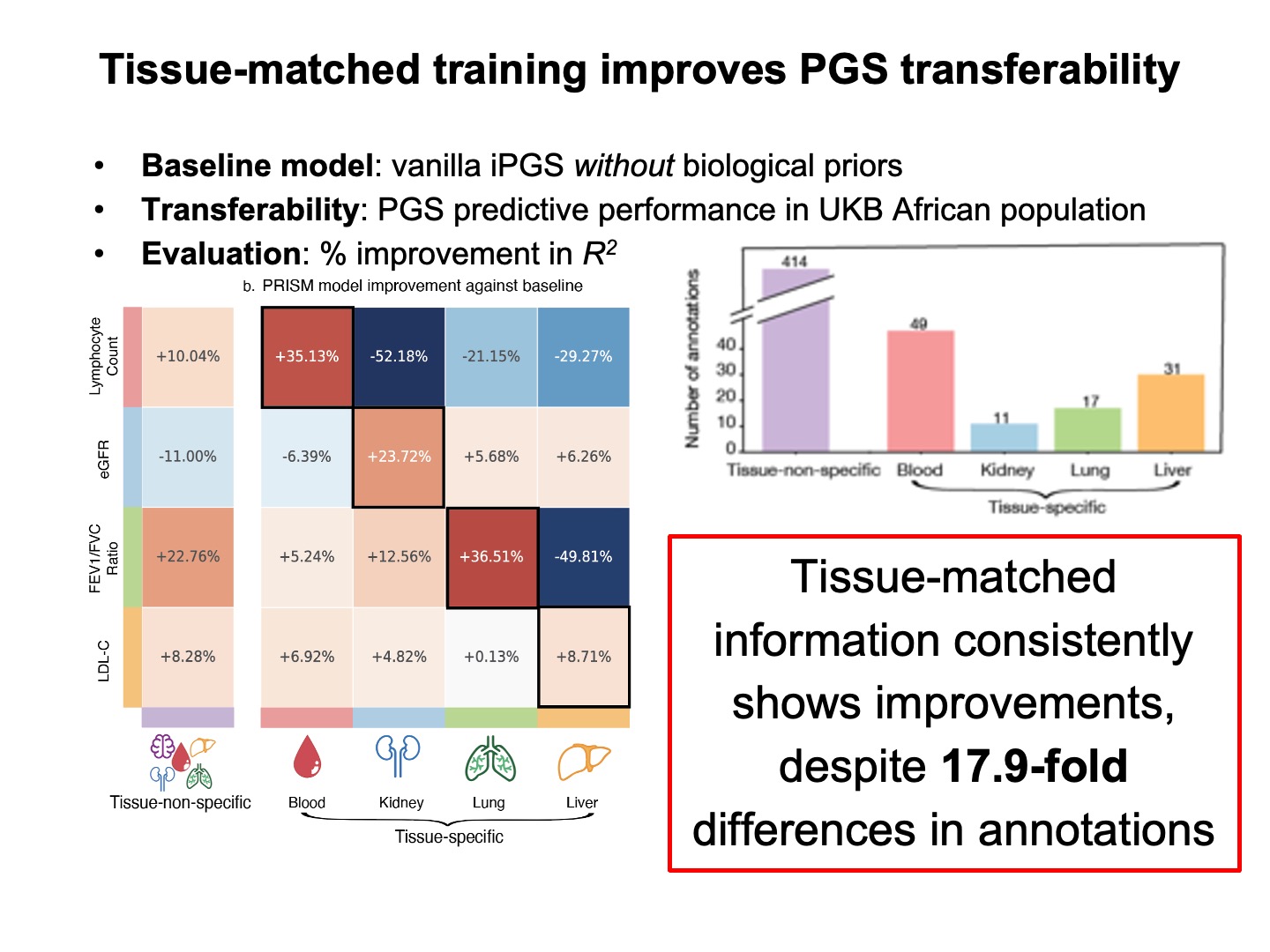
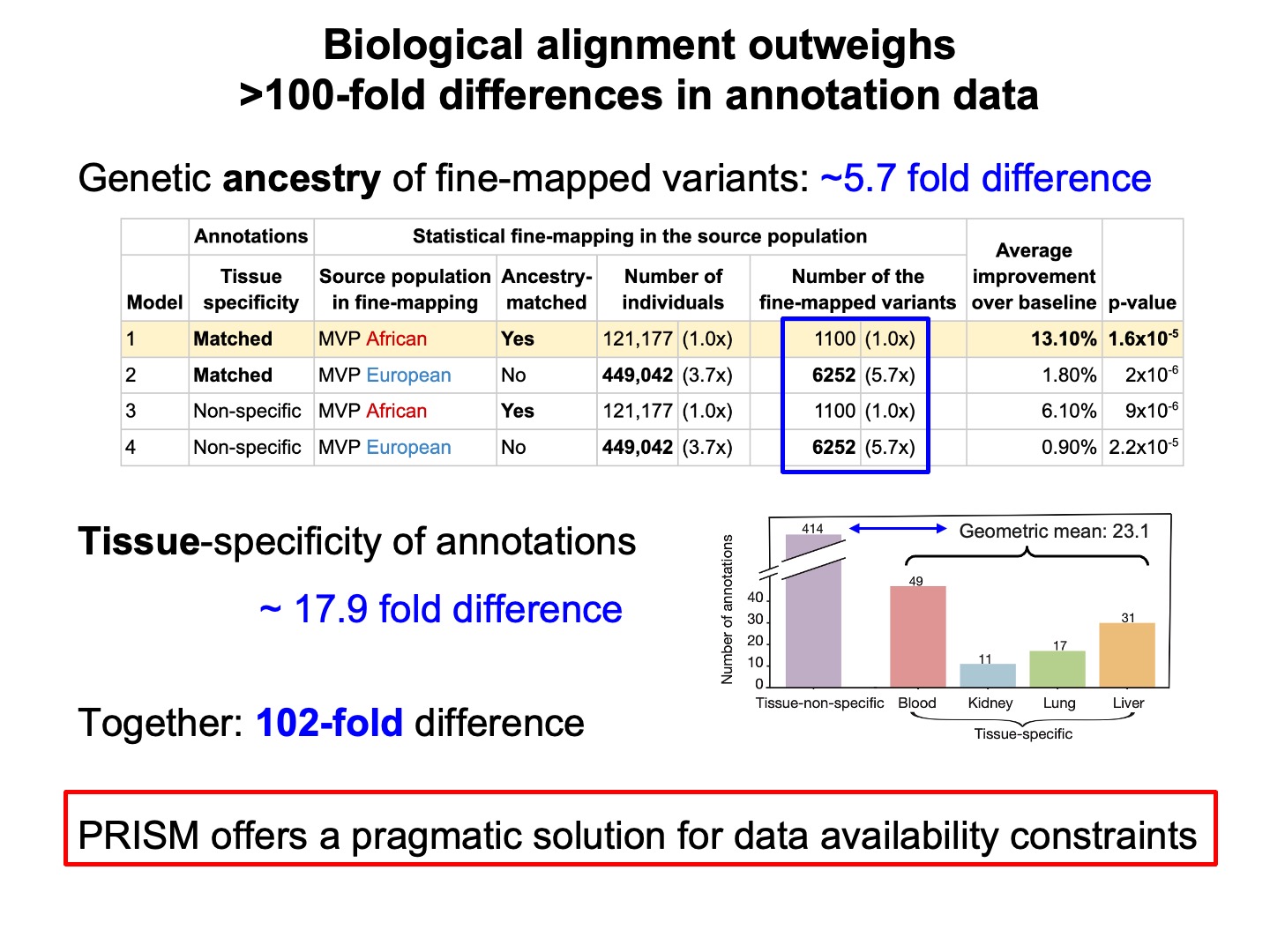
We found that tissue-matched genomic annotations (from ENCODE) are most effective for enhancing PGS transferability, even though the tissue-agnostic strategy can leverage a ~18-fold larger number of genomic annotation datasets.
Similarly, the use of genetic ancestry-specific fine-mapped variants is most effective, despite the power difference. Our approach offers a pragmatic solution for working with data with uneven coverage across contexts.
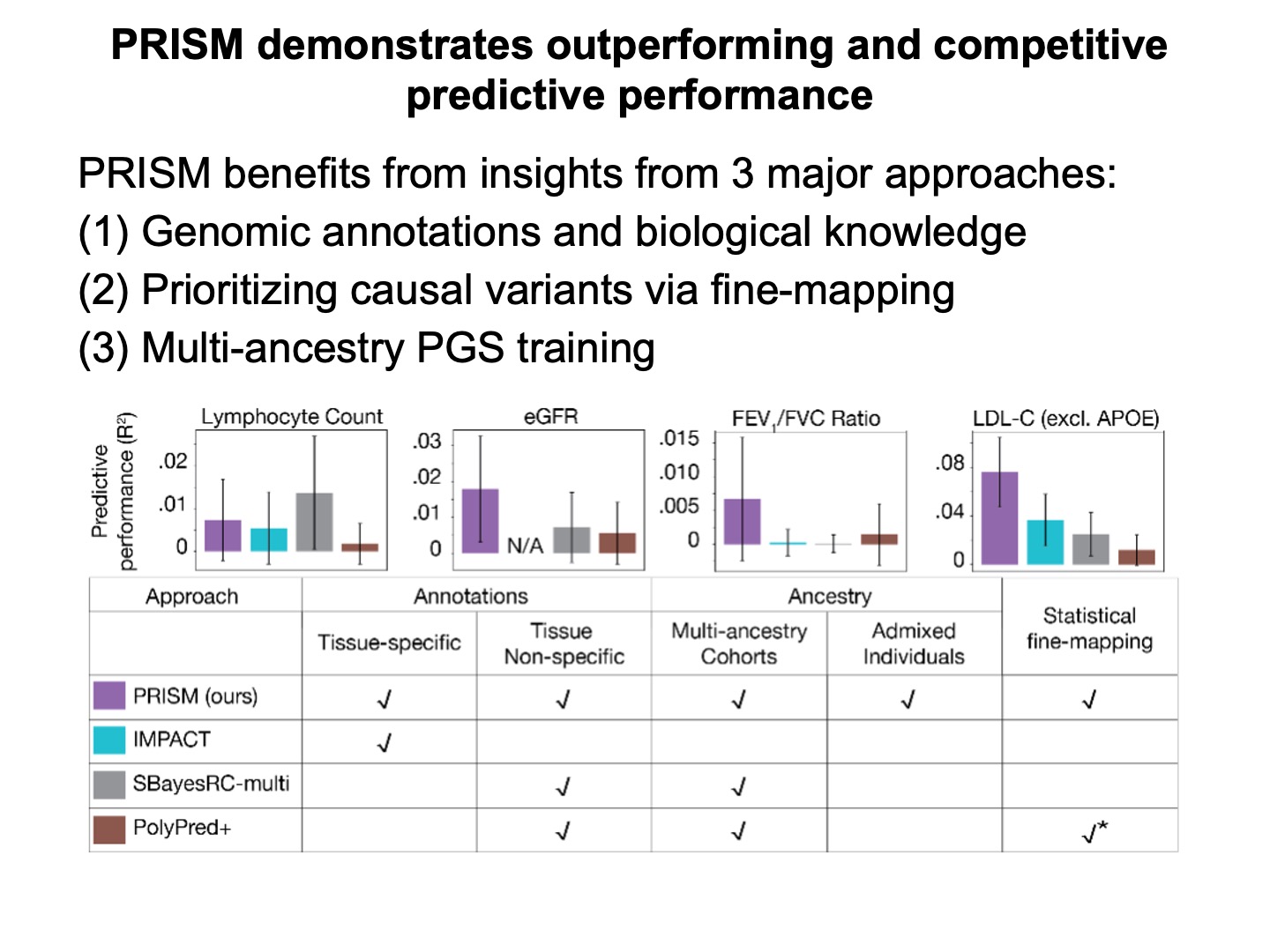
Our benchmarking analysis shows PRISM benefits from three different strategies for enhancing PGS transferability.
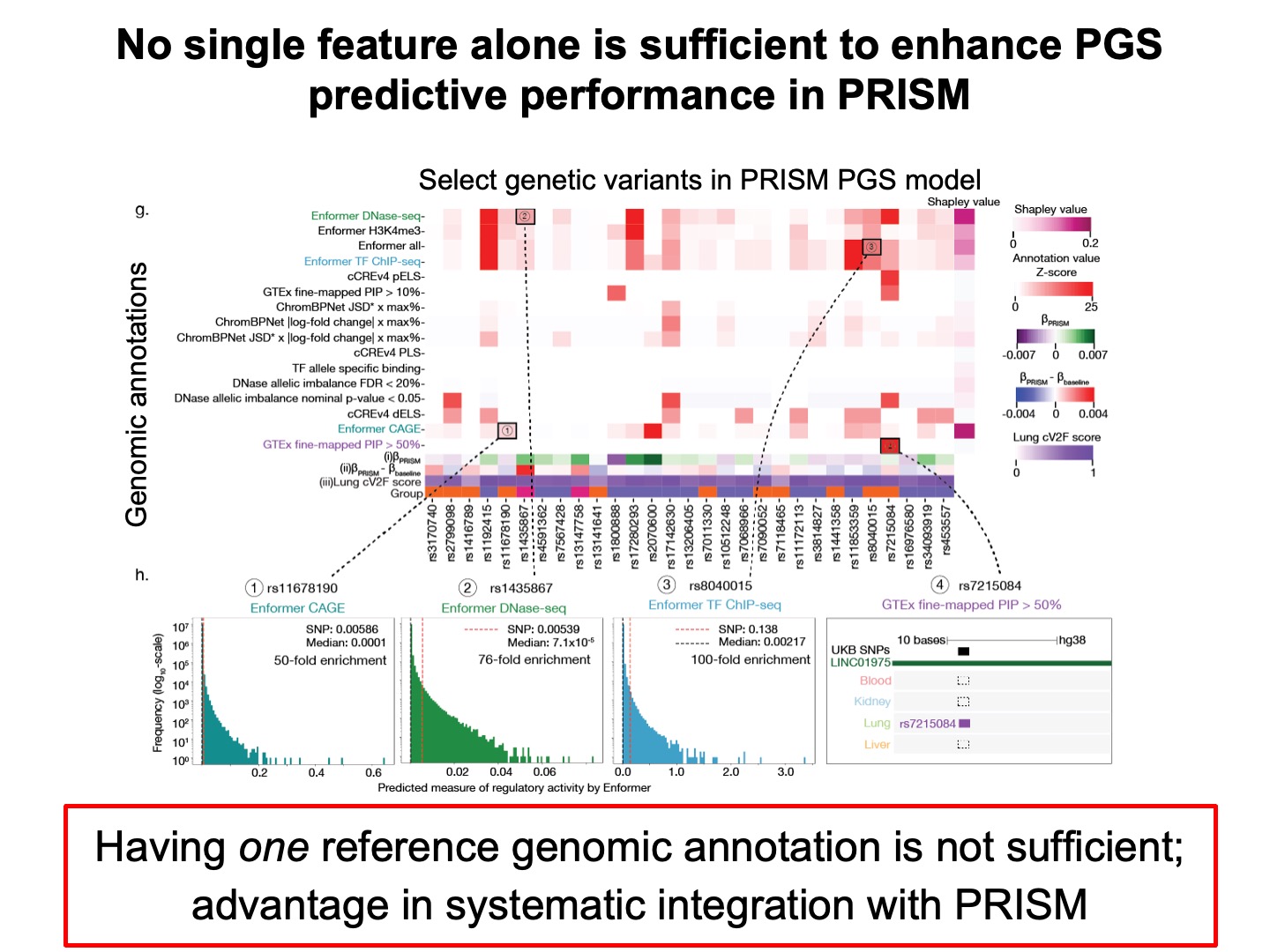
We showed, through feature importance analysis, that having one reference genomic annotation is not sufficient, highlighting the advantage of systematic integration with our approach.
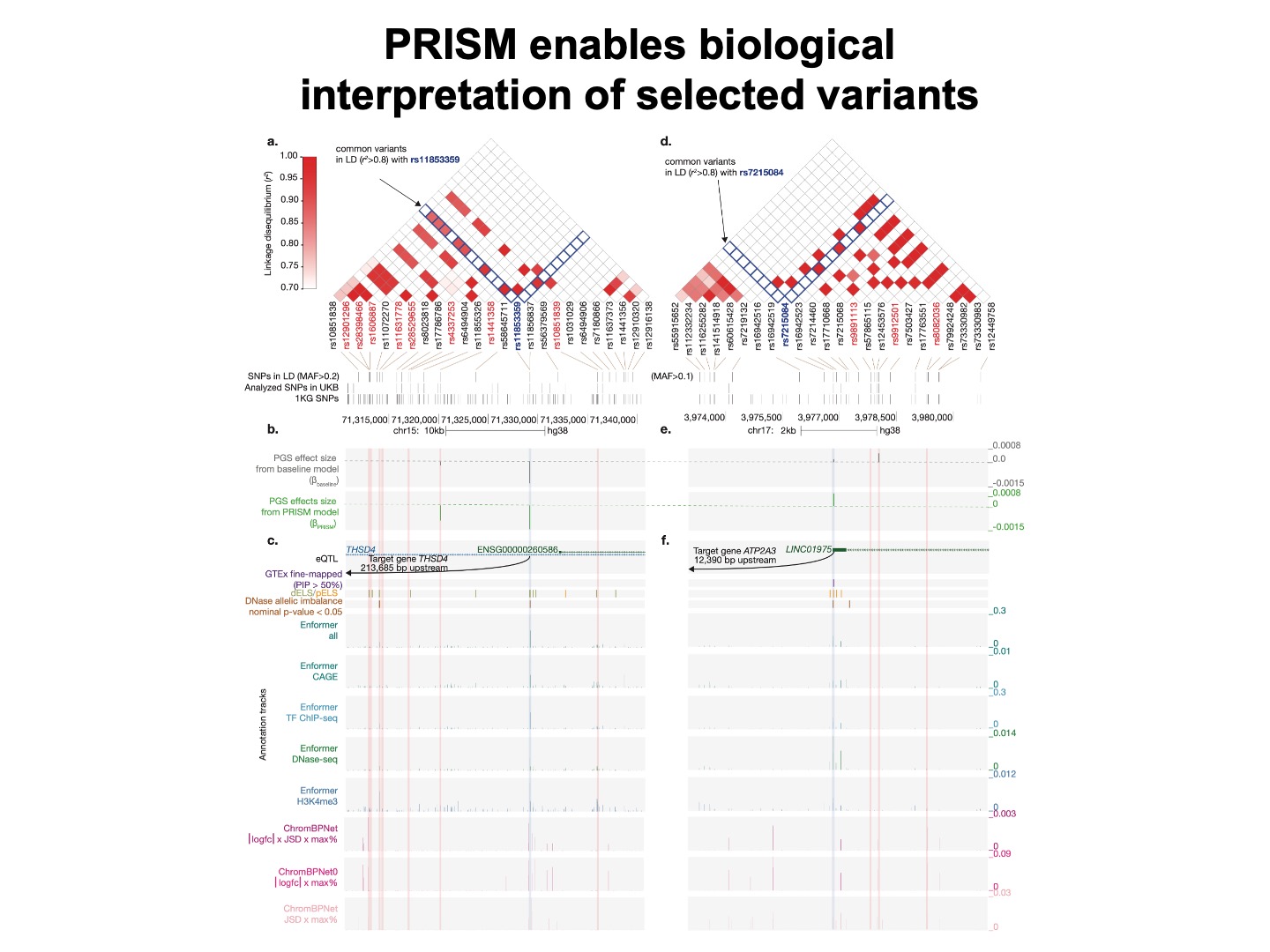
We confirm that the genetic variants selected from our approach are supported by various genomic annotations compared to other variants in LD.
I am grateful to Lucy and the team for making this work possible.
Links
Reference: PRISM: ancestry-aware integration of tissue-specific genomic annotations enhances the transferability of polygenic scores, X. Tian, T. Fabiha, W. F. Li, K. K. Dey, M. Kellis, Y. Tanigawa. bioRxiv, 2025.11.13.688144 (2025). https://doi.org/10.1101/2025.11.13.688144
