Components of genetic associations across 2,138 phenotypes in the UK Biobank highlight adipocyte biology
Published in Nature Communications, 2019
While many pleiotropic genetic loci have been identified, how they contribute to phenotypes across traits and diseases is unclear. We developed DeGAs to address this issue.
Highlights
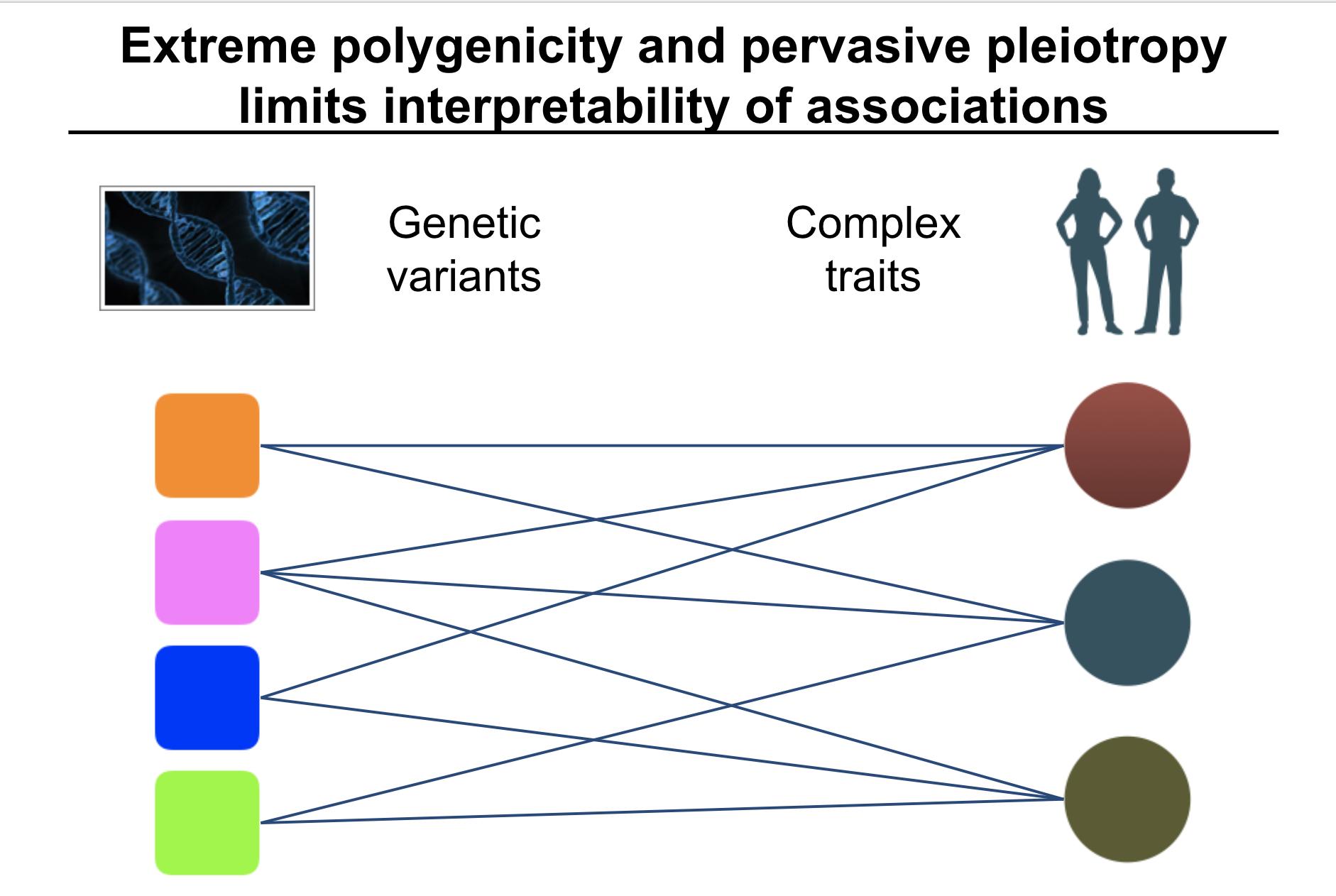
When analyzing the genetics of complex traits, extreme polygenicity and pervasive pleiotropy are challenges in the interpretation and translational application of genetic findings.
To address this challenge, we propose to introduce latent components of genetic associations.
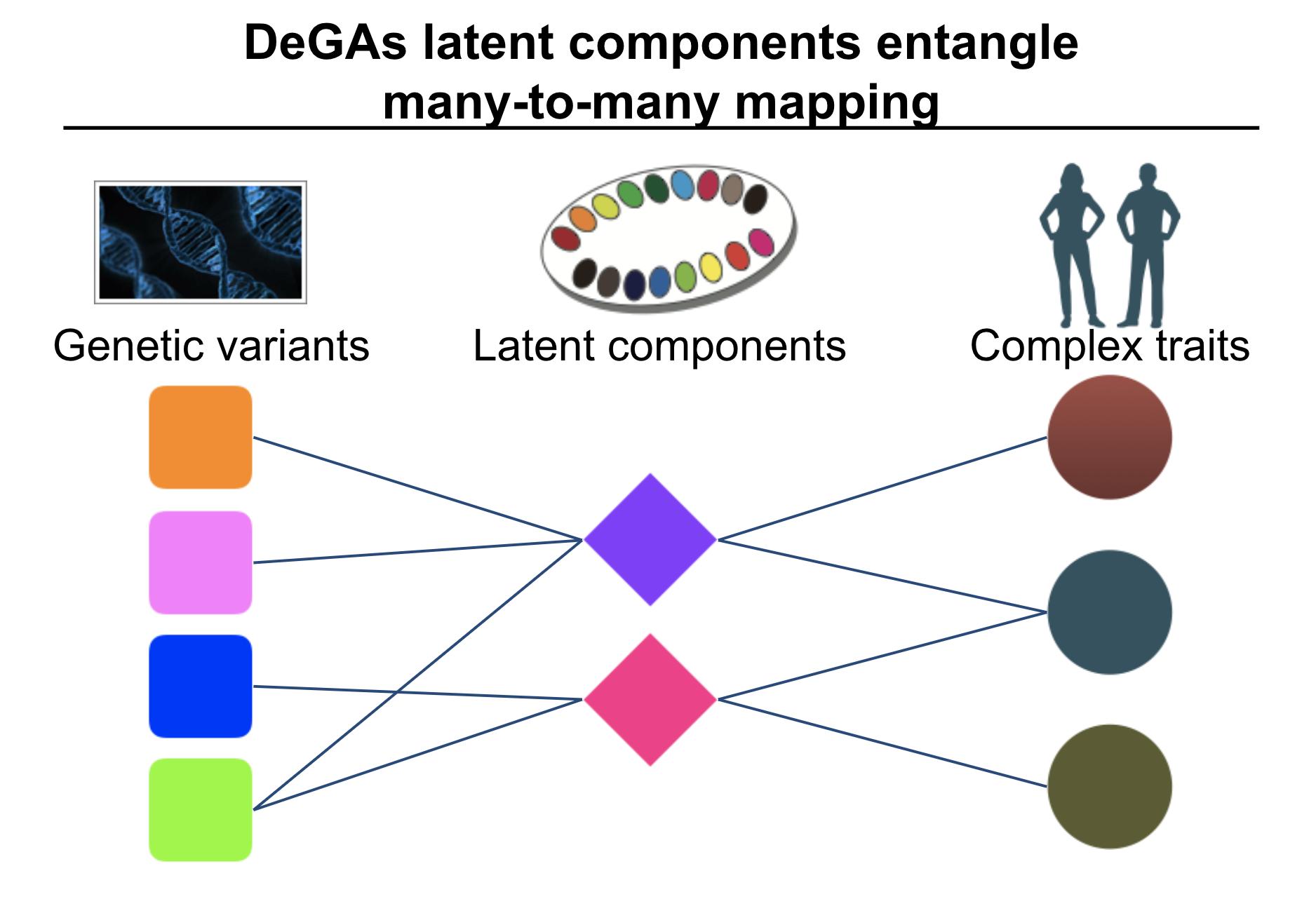
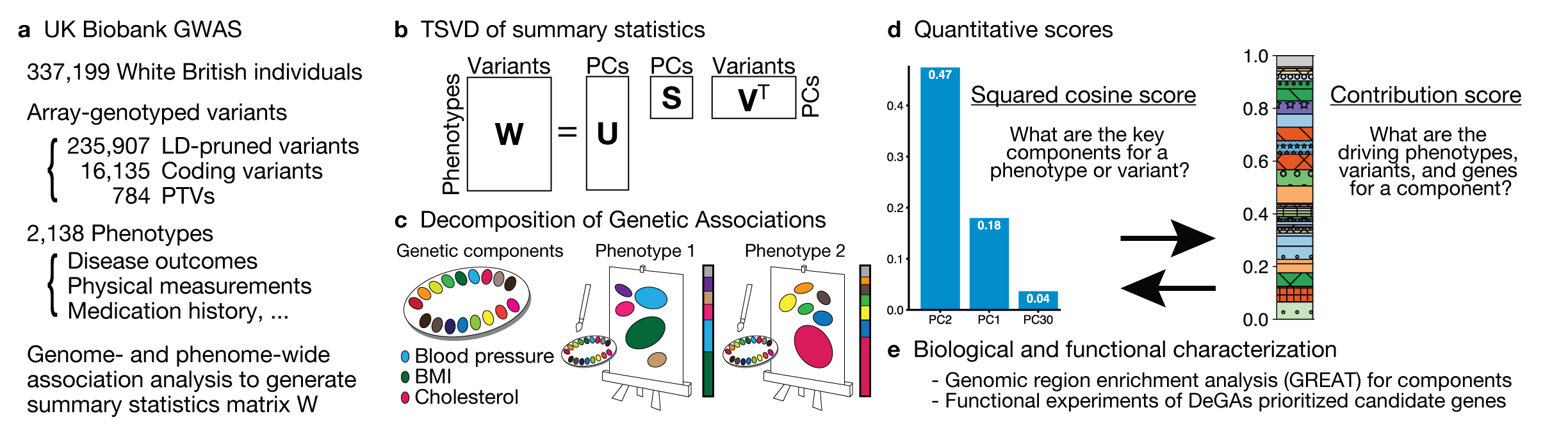
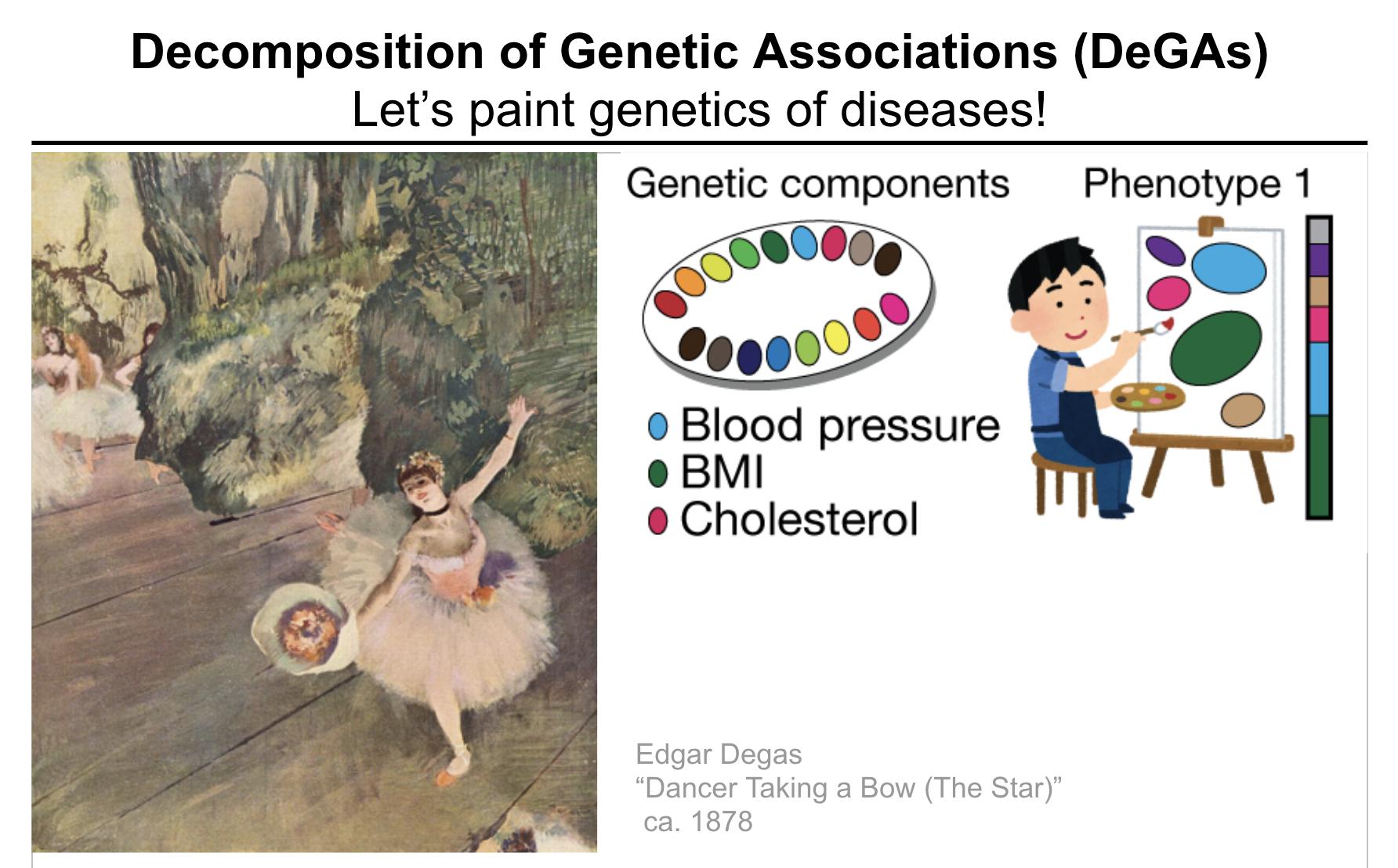
In DeGAs (Decomposition of Genetic Associations), we identify latent components of genetic association by applying truncated singular-value decomposition (TSVD) on a matrix consisting of genome-wide association summary statistics computed for thousands of phenotypes. Using those components and our quantitative scores, we represent the genetics of a disease as a mixture of different components – “painting” of the genetics of diseases. We also characterize components to provide interpretation.
When applied to 2000+ phenotypes in UK Biobank, we found that a related set of phenotypes and variants are captured in DeGAs latent space. For example, standing and sitting heights are in the same direction, even though we applied DeGAs to association summary statistics.
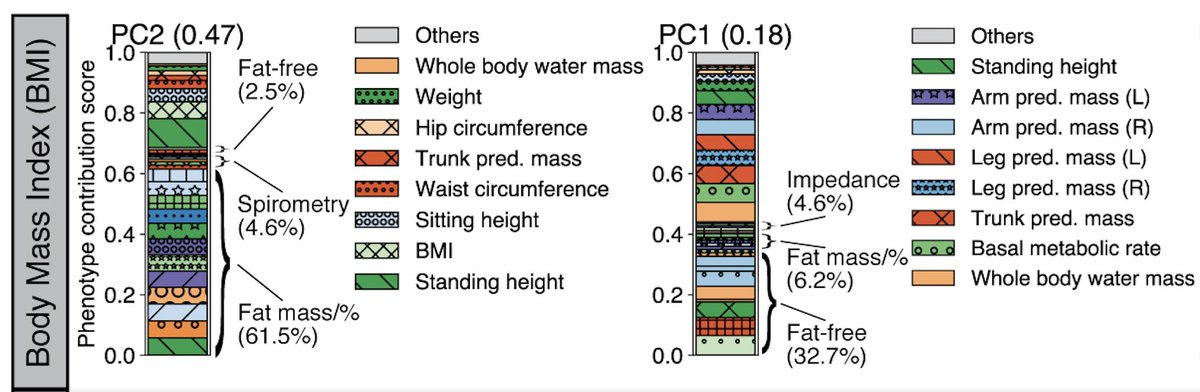
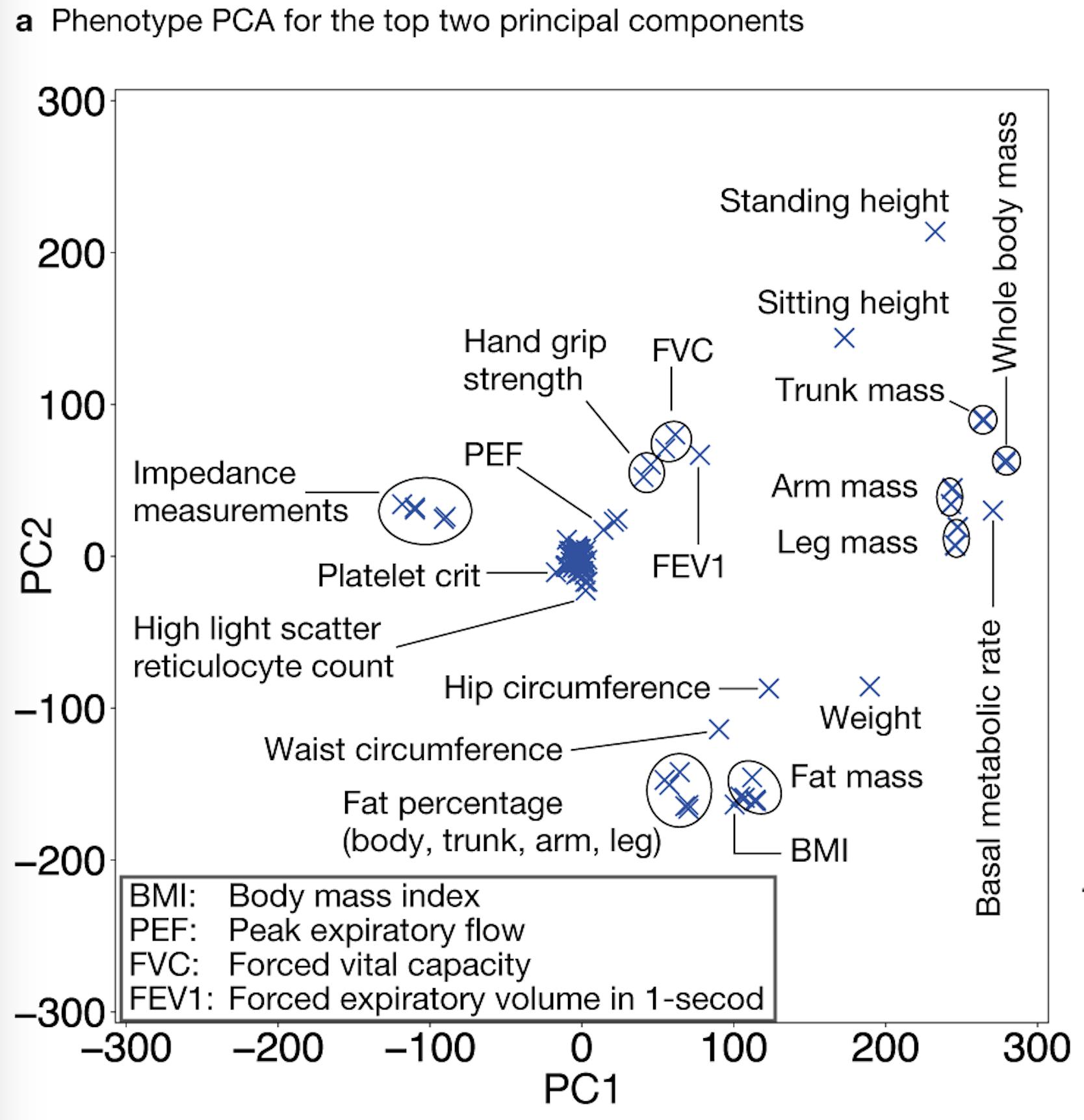 When we look at the top two DeGAs components for body mass index (BMI), the top one (PC2) is mainly driven by fat-related traits, whereas the second most important one (PC1) is mainly driven by fat-free traits, providing an enhanced interpretation of the genetics of BMI.
When we look at the top two DeGAs components for body mass index (BMI), the top one (PC2) is mainly driven by fat-related traits, whereas the second most important one (PC1) is mainly driven by fat-free traits, providing an enhanced interpretation of the genetics of BMI.
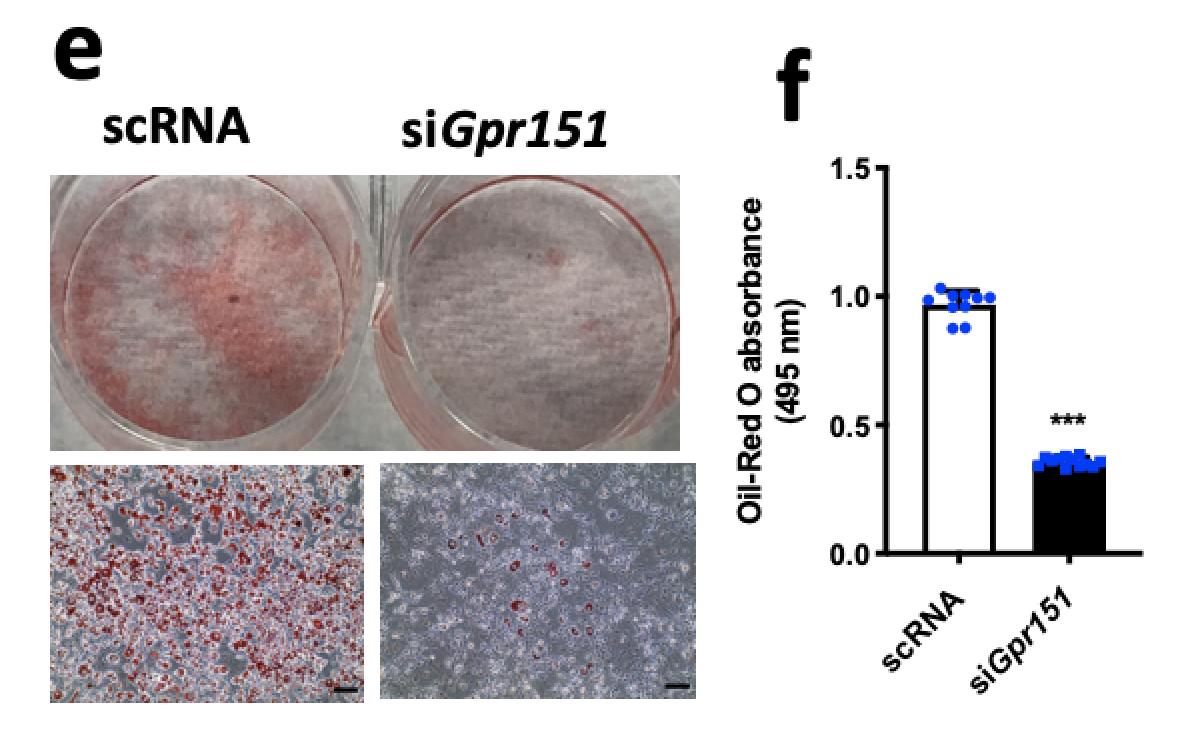
To prioritize genes for experiments, we applied DeGAs to a subset of the dataset consisting of protein-truncating variants and identified PDE3B and GPR151 as the top two candidates for obesity. Our siRNA knockdown of Gpr151 showed a dramatic decrease in lipids in adipocytes!
Some extensions of DeGAs
In the Rivas lab, we have several projects that extend the work presented in DeGAs.
- DeGAs-PRS (dPRS): We propose
dPRS, a method to enhance the interpretability of polygenic risk score (PRS) using DeGAs latent components. - Sparse reduced-rank regression (SRRR): In DeGAs, we took the summary statistics from univariate association scan across genetic variants and phenotypes. We propose a method to directly fit multi-response sparse regression models.
Resource
We provide a resource for the research community. We developed interactive DeGAs web application as a part of Global Biobank Engine, whose video tutorial is shown above.
The datasets used in the study are available from figshare.
Y. Tanigawa, and M. A. Rivas, Decomposed matrices used for the analysis described in ‘Components of genetic associations across 2,138 phenotypes in the UK Biobank highlight adipocyte biology’. https://doi.org/10.35092/yhjc.9202247.v1 (2019).
Coverage
- Barroso I and Wheeler E: Faculty Opinions Recommendation of [Tanigawa Y et al., Nat Commun 2019 10(1):4064]. In Faculty Opinions, 18 Sep 2019; doi:10.3410/f.736565849.793565082.
- Stanford Cardiovascular Institute Articles & Announcements, A method to shed light on the genetic underpinnings of complex human traits
Reference: Y. Tanigawa*, J. Li*, J. M. Justesen, H. Horn, M. Aguirre, C. DeBoever, C. Chang, B. Narasimhan, K. Lage, T. Hastie, C. Y. Park, G. Bejerano, E. Ingelsson, M. A. Rivas, Components of genetic associations across 2,138 phenotypes in the UK Biobank highlight adipocyte biology. Nat Commun. 10, 4064 (2019). https://doi.org/10.1038/s41467-019-11953-9
