Fast Lasso method for large-scale and ultrahigh-dimensional Cox model with applications to UK Biobank
Published in Biostatistics, 2020
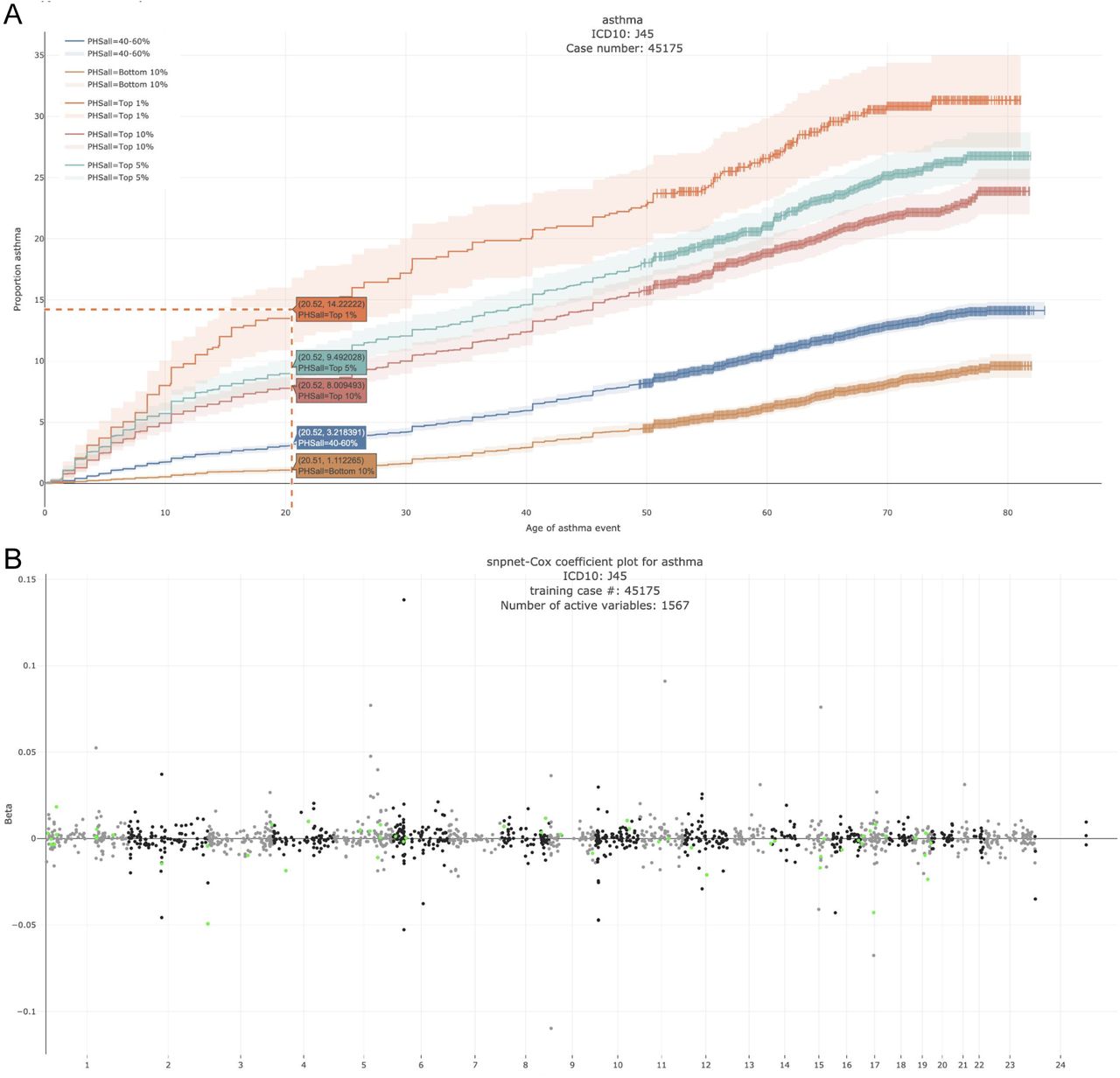
In this study led by Ruilin Li, we propose extending the BASIL/snpnet algorithm to fit the L1 penalized Cox proportional hazards model using a large-scale dataset from a genotyped cohort. We present its application to 300+ time-to-event traits in UK Biobank.
Note that this project is a Cox-model extension of BASIL algorithm. Please check the BASIL/snpnet paper for more in-depth discussion on the BASIL algorithm and snpnet package.
Datasets
We provide the datasets related to this study.
polygenic hazard score
We provide the coefficients of the polygenic hazard score computed for the 306 time-to-event phenotypes in UK Biobank.
- R. Li, C. Chang, J. Justesen, Y. Tanigawa, J. Qian, T. J. Hastie, M. Rivas, R. Tibshirani, snpnet-Cox results (2020), doi:10.6084/m9.figshare.12368294.v1.
Reference: R. Li, C. Chang, J. M. Justesen, Y. Tanigawa, J. Qiang, T. Hastie, M. A. Rivas, R. Tibshirani, Fast Lasso method for large-scale and ultrahigh-dimensional Cox model with applications to UK Biobank. Biostatistics. 23(2), 522-540 (2020). https://doi.org/doi:10.1093/biostatistics/kxaa038
