Significant Sparse Polygenic Risk Scores across 813 traits in UK Biobank
Published in PLOS Genetics, 2022
Using the dense phenotypic information in UK Biobank, we systematically characterized polygenic risk score (or PRS) across more than 1,500 traits. We evaluated the predictive performance of the models and compared that with the baseline models that only consider covariates, which are age, sex, and top 10 genotype principal components. We then assessed the statistical significance of the PRS in improving the predictive performance. When we look at the incremental R2 or incremental AUC, we find 813 traits have significant incremental predictive performance, after accounting for multiple hypothesis testing.
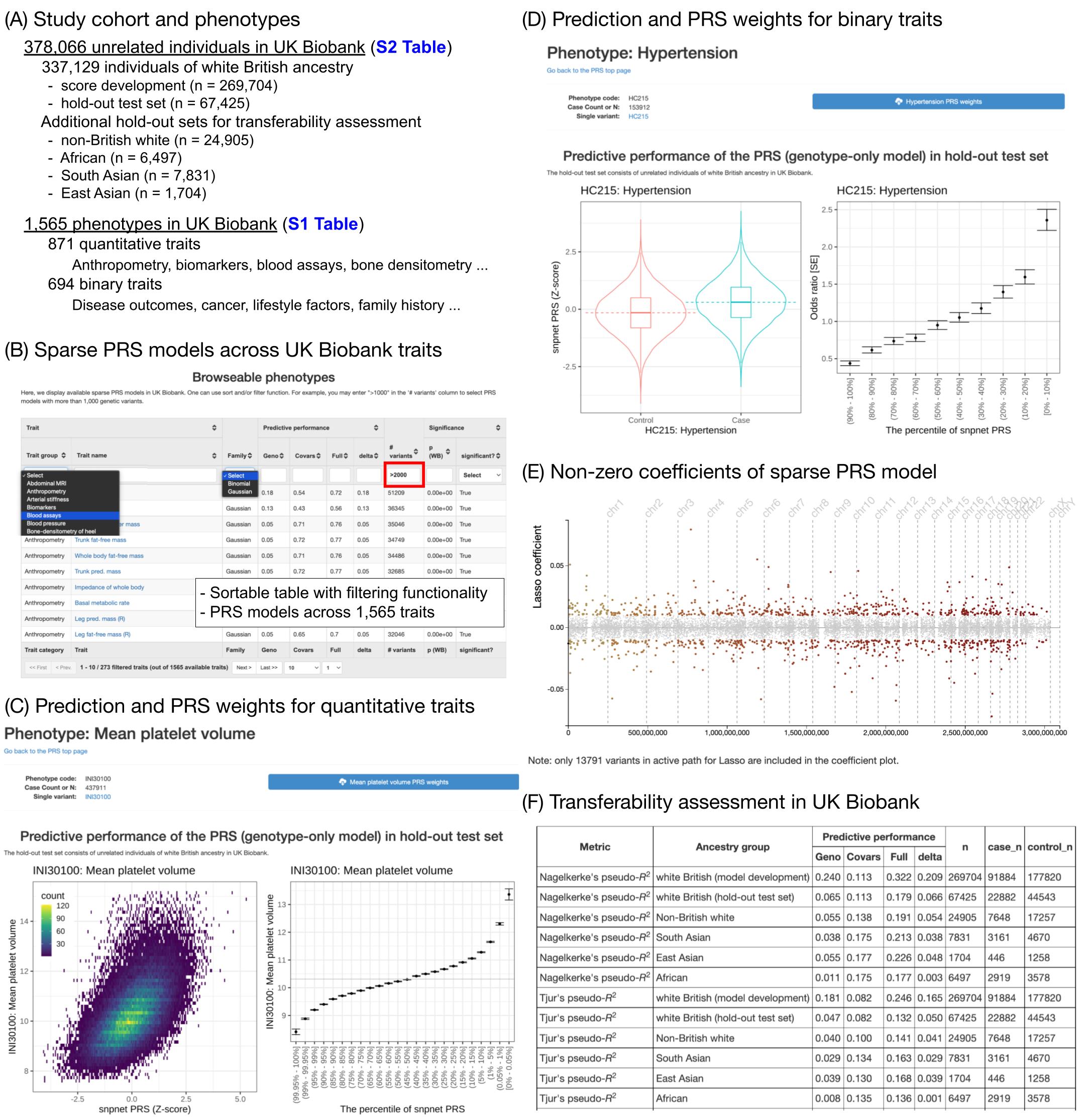
Across 813 traits with significant PRS, we asked whether the number of genetic variants in the sparse PRS models is correlated with the incremental predictive performance. We found those two quantities are significantly correlated in both quantitative traits and binary traits.
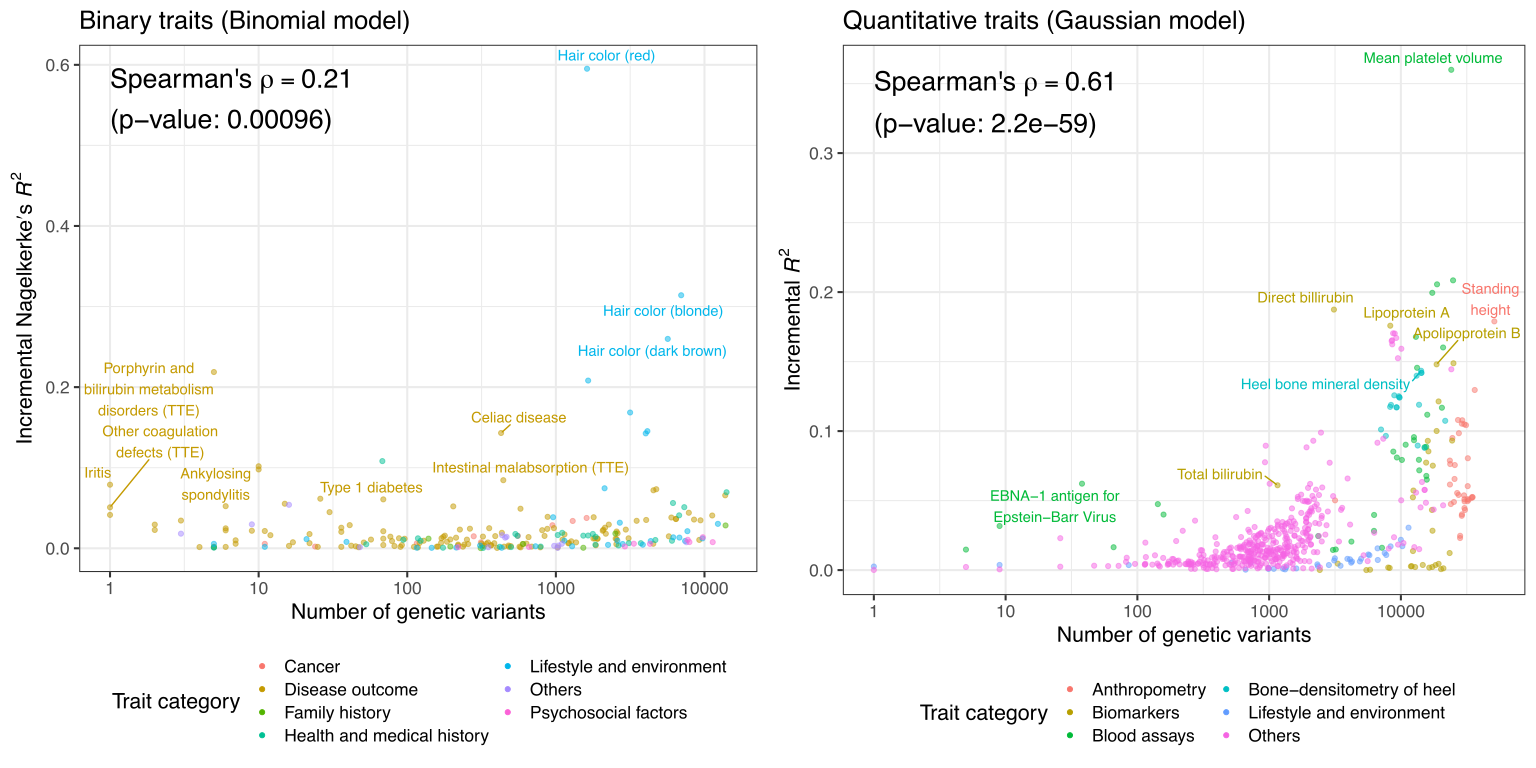
We assessed the trans-ethnic predictive performance using the additional test sets of different ancestry groups in UK Biobank.
If you’re interested, please look at our PRS map page at the Global Biobank Engine. If you’re interested in the BASIL algorithm and the R snpnet package that we used in this application, please also look at our paper (J. Qian, Tanigawa, et al, 2020)
Revision history
2022.03.24
Our paper is published at PLOS genetics. We thank insightful feedback from the reviewers.
2022.01.27
We revised the manuscript based on the feedback from colleagues. The major changes in this revision are the following three points.
- Given the feedback from colleagues, we removed the sentences that inappropriately mentioned genetic architecture in binary traits. We instead clarified a power difference between quantitative traits and binary traits.
- Given the concerns regarding the lack of theoretical basis in using incremental ROC-AUC for assessing linear relationship (estimated SNP-based heritabilities and transferability assessment), we now use Nagelkerke’s pseudo-R2 as the primary evaluation metric of predictive performance for binary traits in the current version of the manuscript.
- As we change the evaluation metric for binary traits, we now observe a significant rank-based correlation between the effect size (incremental Nagelkerke’s pseudo-R2) and the model size (number of genetic variants with non-zero coefficients) of the sparse PRS model.
2021.11.26
We have updated the manuscript based on the feedback from the colleagues. The major changes in this revision are the following six points.
- We reported the significant polygenic risk score (PRS) models for 428 traits in the original manuscript. We realized that there was a critical mistake when evaluating the significance of the PRS models (specifically, the coefficients of covariates were mistakenly estimated on the test set individuals, not on the score development set). We fixed this issue and now present 813 significant PRS models in the latest version of the manuscript.
- Following the feedback from colleagues, we included the types of genotyping array in the covariates when evaluating the predictive performance of PRS models. We updated the predictive performance metrics.
- We added Table 1 and Supplementary Table 3 and described the effects of prioritization of medically relevant alleles.
- We added a new figure (Fig 2) comparing the estimated SNP-based heritability vs. the predictive performance of PRS models.
- We deposit the significant PRS models on the PGS catalog. We list the score IDs in Supplementary Table 1.
- We improved the clarity of the methods section and added a few additional analyses (Supplementary Figure 1 - Supplementary Figure 3).
2021.09.02
We posted the original version on medRxiv.
Data and Code availability
- The sparse PRS model weights generated from this study are available on the Global Biobank Engine.
- The significant PRS models are also available at the PGS catalog (PGP000244 and PGP000128, score IDs are listed in S1 Table).
- The BASIL algorithm implemented in the R snpnet package was used in the PRS analysis.
- The analyses presented in this study were based on data accessed through the UK Biobank
日本語での解説 (Highlights in Japanese)
近年、ポリジェニック・スコア(PGS)と呼ばれる統計解析技術と、その臨床応用への関心が高まっている。ポリジェニック・スコアは、各個人のゲノムに含まれる数千〜数百万もの遺伝子変異の効果を足し合わせ、個人レベルの疾患リスクや疾患関連形質を予測を行う。アメリカ心臓協会が科学的声明を発表するなどPGSの臨床応用への期待も高まっている。心臓病・肺がんなど、遺伝学研究が充実している疾患領域でのPGS研究は先行している。一方、電子カルテやコホート型研究により得られるヒトの幅広い諸形質に対して、PGSを用いることが有用であるかは明らかにされていなかった。
本論文では、英国 UK Biobank の約378,000人のデータを用いてポリジェニック・スコア・モデルを構築し、その網羅的な性能評価を行った。わたしたちは、871の連続値の形質と694の二値形質、合計1565の形質情報を整備した。連続値をとる形質には、血液バイオマーカーや身長に加え脳MRI画像を処理して得られる形態学的情報などが含まれ、二値形質には、疾患の症例・対照例や生活習慣などが含まれる。これらに対してポリジェニック・スコア・モデルの有用性を網羅的に評価し、813 形質において統計学的に有意な予測精度を示すポリジェニック・スコア・モデルが得られることを示した。また、ポリジェニック・スコアモデルの予測精度は、各形質の集団中での分布がどの程度遺伝要因により影響を受ける程度を表す遺伝率による制約をうけること、予測性能が高いポリジェニック・スコア・モデルには、より多くの遺伝子変異の項が含まれることを経験的に示した。
得られた813形質に対するポリジェニック・スコア・モデルは、ウェブサイトにて公開されている。
Reference: Y. Tanigawa, J. Qian, G. R. Venkataraman, J. M. Justesen, R. Li, R. Tibshirani, T. Hastie, M. A. Rivas, Significant Sparse Polygenic Risk Scores across 813 traits in UK Biobank. PLOS Genet. 18(3), e1010105 (2022). https://doi.org/10.1371/journal.pgen.1010105
