Power of inclusion: Enhancing polygenic prediction with admixed individuals
Published in The American Journal of Human Genetics, 2023
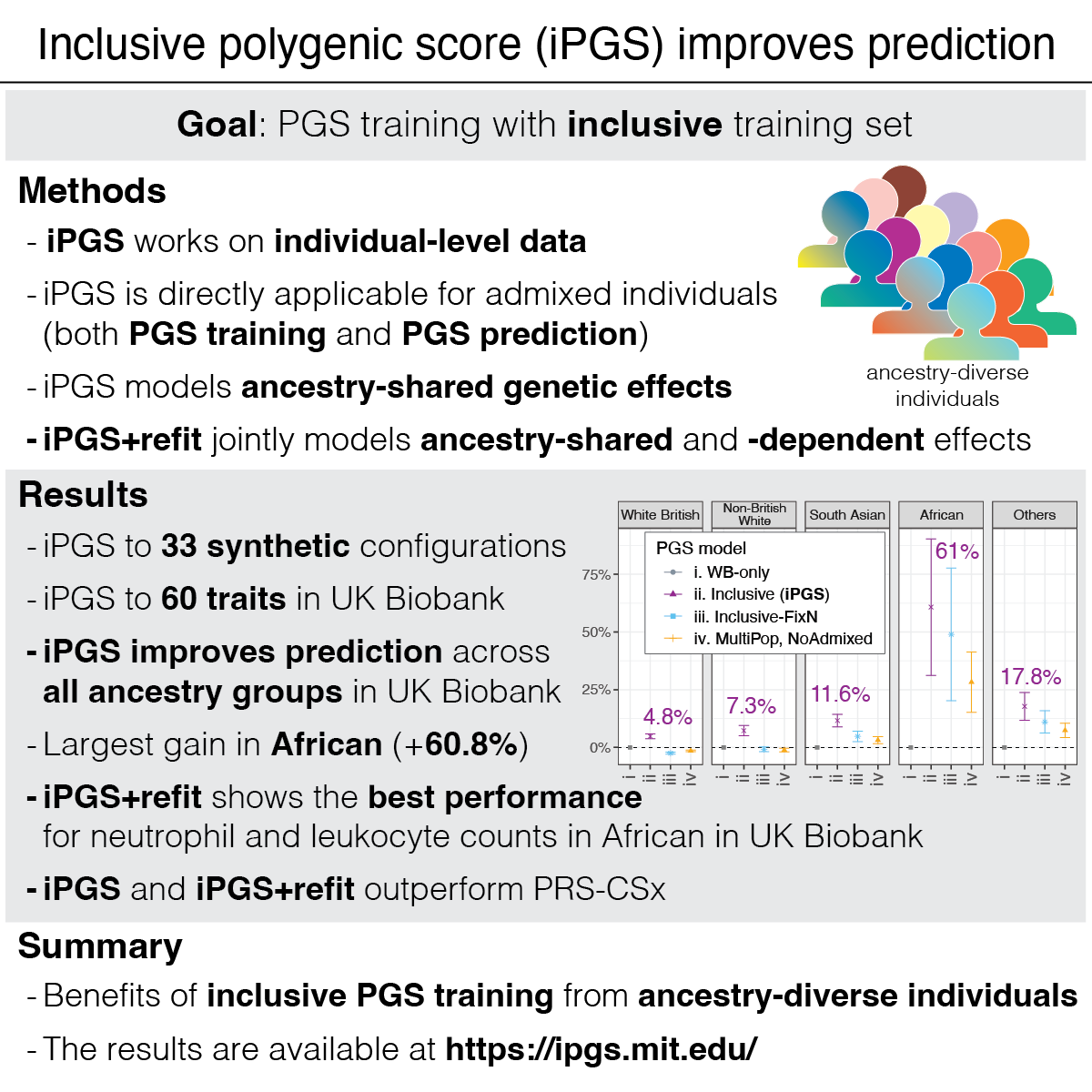
We developed a polygenic score training approach that allows direct inclusion of admixed individuals without the need for local ancestry inference and showed ancestry-diverse training improves prediction for all tested population groups. The MIT News article provides a fantastic summary of our work.
Abstract
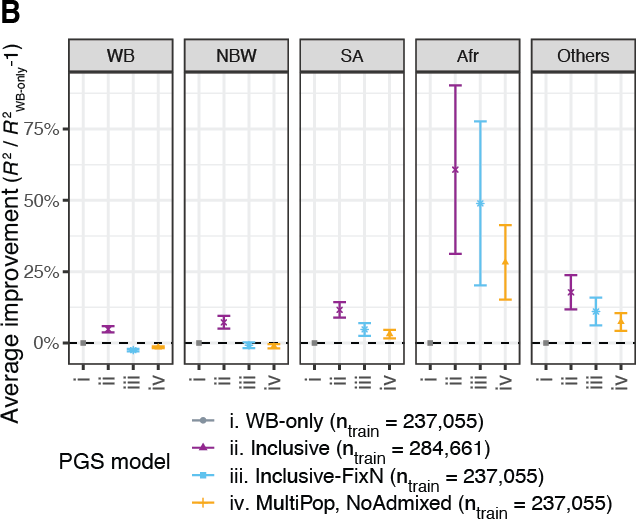
Admixed individuals offer unique opportunities for addressing limited transferability in polygenic scores (PGSs), given the substantial trans-ancestry genetic correlation in many complex traits. However, they are rarely considered in PGS training, given the challenges in representing ancestry-matched linkage-disequilibrium reference panels for admixed individuals. Here we present inclusive PGS (iPGS), which captures ancestry-shared genetic effects by finding the exact solution for penalized regression on individual-level data and is thus naturally applicable to admixed individuals. We validate our approach in a simulation study across 33 configurations with varying heritability, polygenicity, and ancestry composition in the training set. When iPGS is applied to n = 237,055 ancestry-diverse individuals in the UK Biobank, it shows the greatest improvements in Africans by 48.9% on average across 60 quantitative traits and up to 50-fold improvements for some traits (neutrophil count, R2 = 0.058) over the baseline model trained on the same number of European individuals. When we allowed iPGS to use n = 284,661 individuals, we observed an average improvement of 60.8% for African, 11.6% for South Asian, 7.3% for non-British White, 4.8% for White British, and 17.8% for the other individuals. We further developed iPGS+refit to jointly model the ancestry-shared and -dependent genetic effects when heterogeneous genetic associations were present. For neutrophil count, for example, iPGS+refit showed the highest predictive performance in the African group (R2 = 0.115), which exceeds the best predictive performance for the White British group (R2 = 0.090 in the iPGS model), even though only 1.49% of individuals used in the iPGS training are of African ancestry. Our results indicate the power of including diverse individuals for developing more equitable PGS models.
Highlights
Polygenic score (PGS), a statistical approach to estimating genetic predisposition on traits, attracted substantial research interest. The current PGS models show limited transferability across populations, and there are a number of great new methods to address this challenge.
We propose inclusive polygenic score (iPGS), a PGS training strategy to capture ancestry-shared genetic effects by analyzing individuals across the continuum of genetic ancestry. We work directly on the individual-level data without relying on GWAS results and LD references.
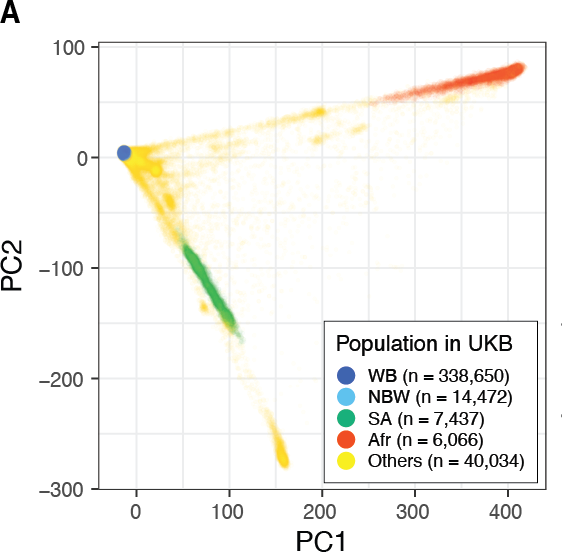
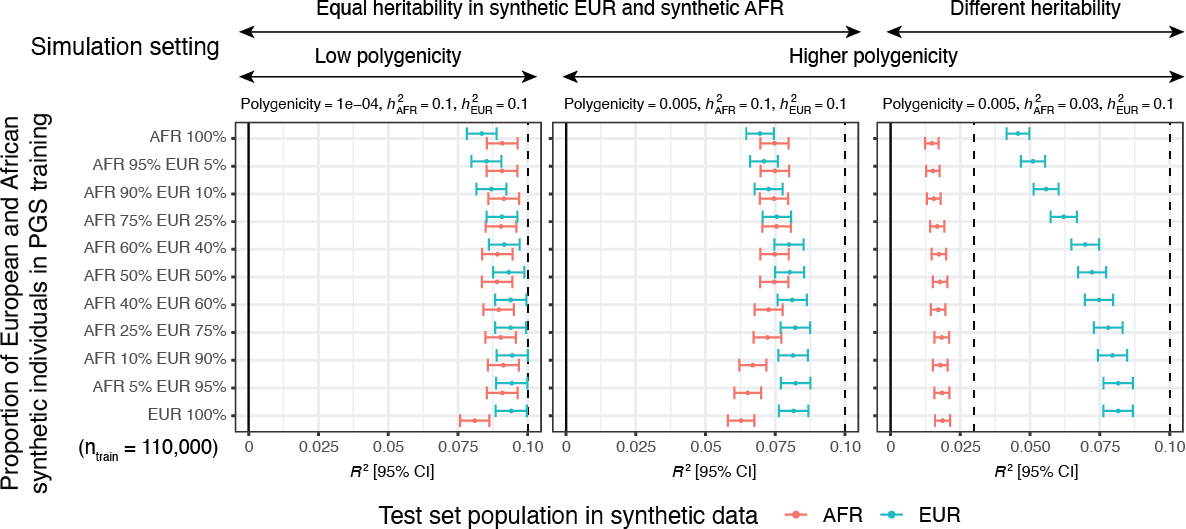
We tested our approach across 33 simulation configurations and 60 quantitative traits in UK Biobank. We see increased power by including ancestry-diverse individuals compared to our baseline model trained only on white British individuals.
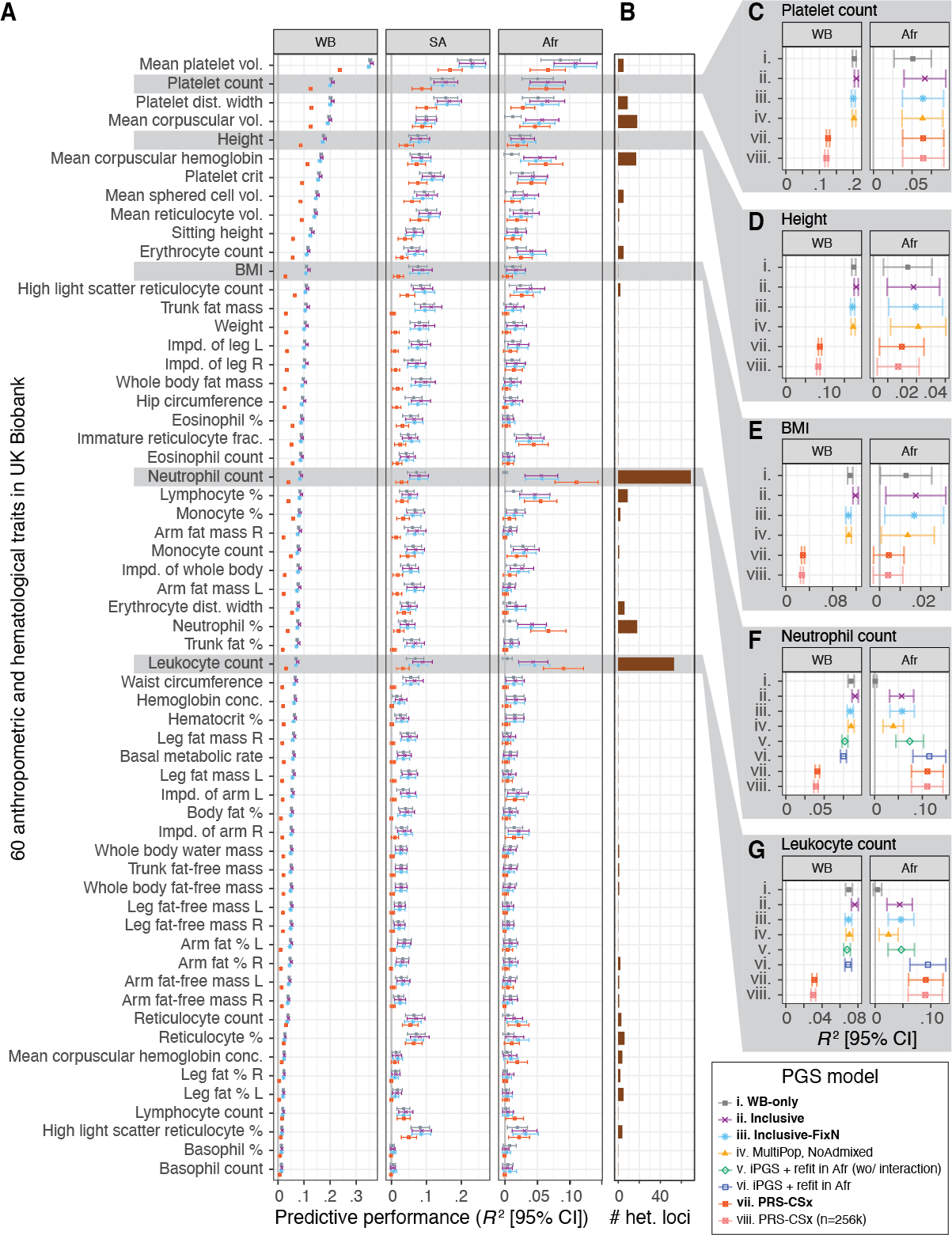
We observe improvements in performance for all population groups. The average improvement across the 60 traits was 60.7% for African, 11.6% for South Asian, 7.3% for non-British white, 4.8% for White British, and 17.8% for other diverse individuals.
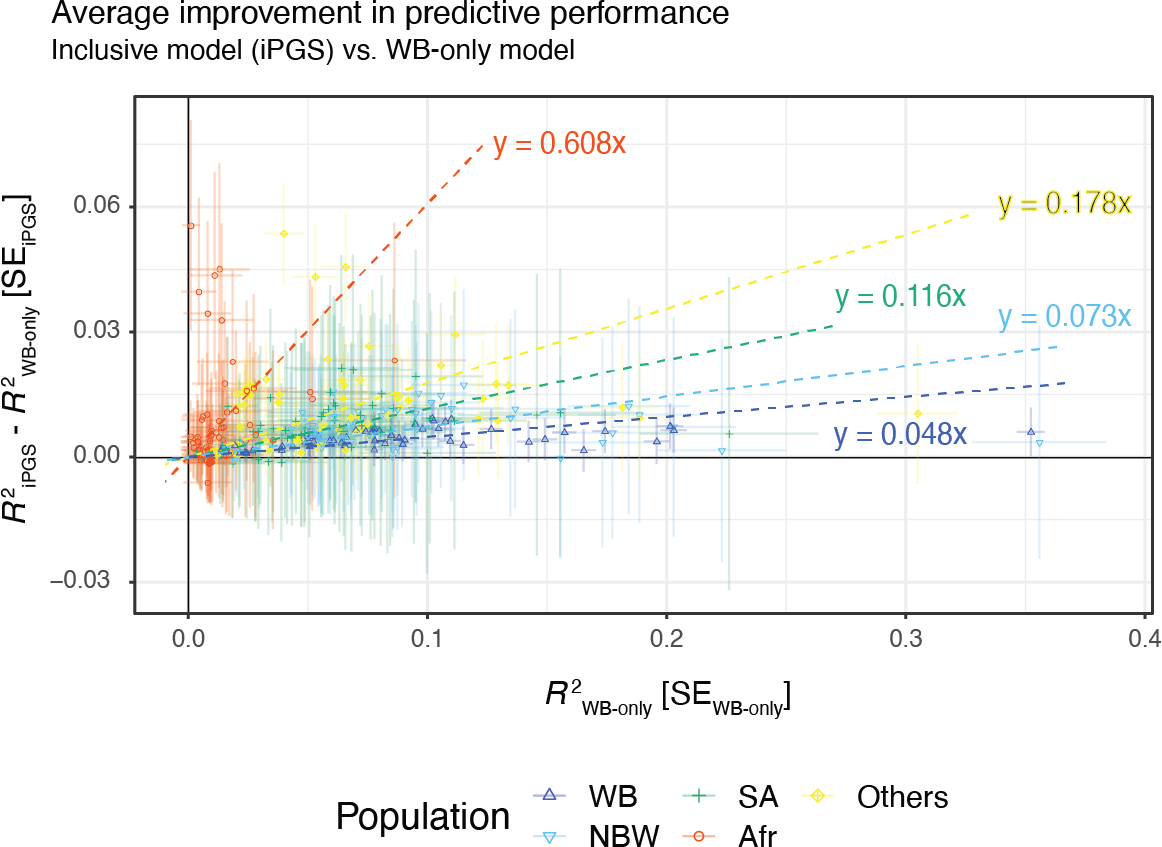
To consider ancestry-dependent genetic effects on top of ancestry-shared effects, we developed iPGS+refit. We used a heterogeneity test in GWAS meta-analysis and identified genetic variants with heterogeneous associations, such as the ACKR1 locus for neutrophil count.
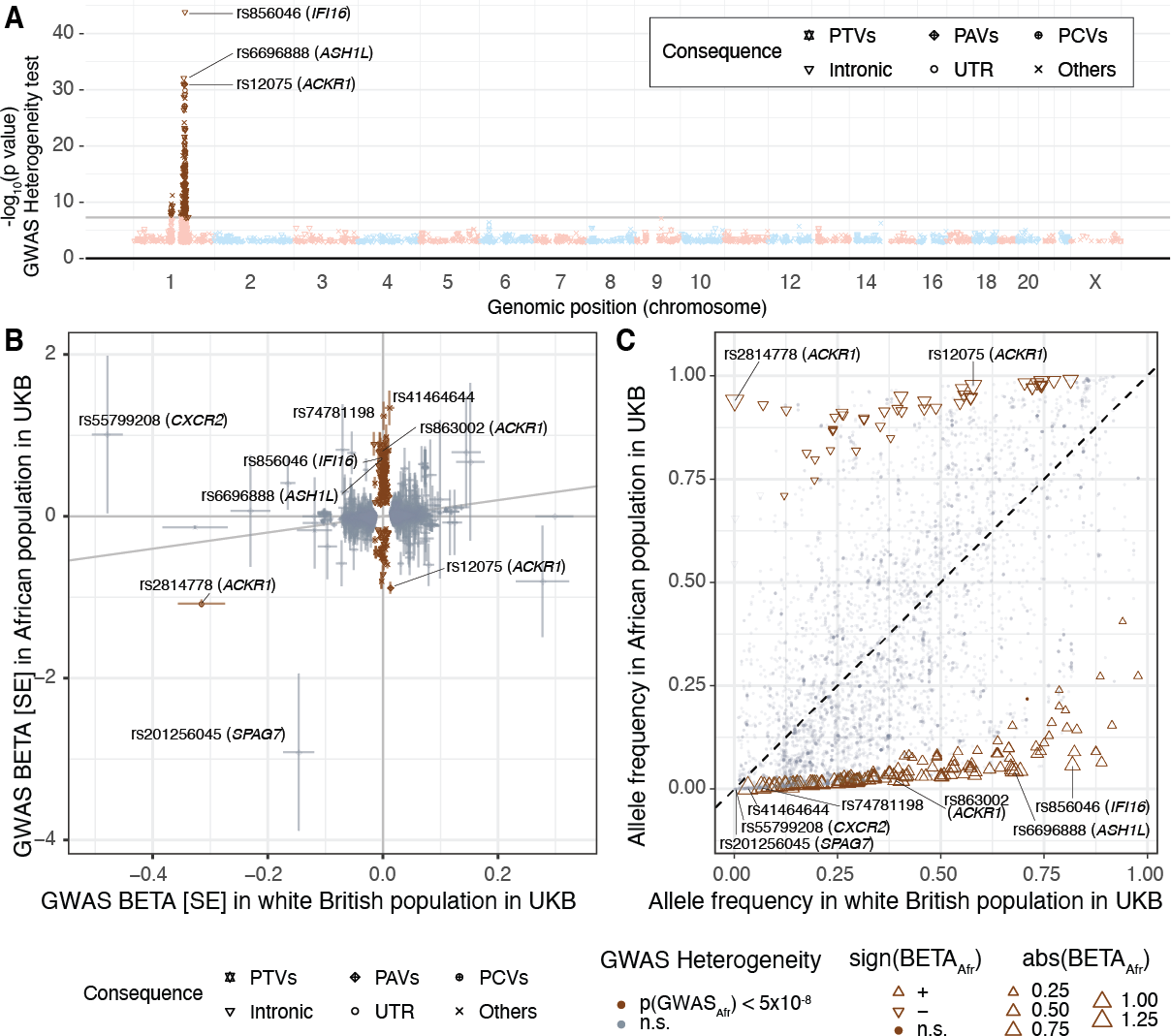
Our iPGS+refit starts with one ancestry-shared component (iPGS) and adds ancestry-dependent effects using a relatively small number of genetic loci, facilitating better interpretation. We used hematological traits to show improved predictive performance.
We compared our model with PRS-CSx, a commonly-used multi-ancestry PGS method from summary statistics from multiple population groups and ancestry-matched reference panels. In our analysis, our iPGS/iPGS+refit models showed competitive or improved performance.
We thank UK Biobank, its participants, amazing collaborators and colleagues, as well as funding.
The iPGS browser and dataset availability
You can browse and download our iPGS models at our iPGS browser. Taking advantage of the sparsity of our PGS models, it offers direct integration with HaploReg and GREAT.
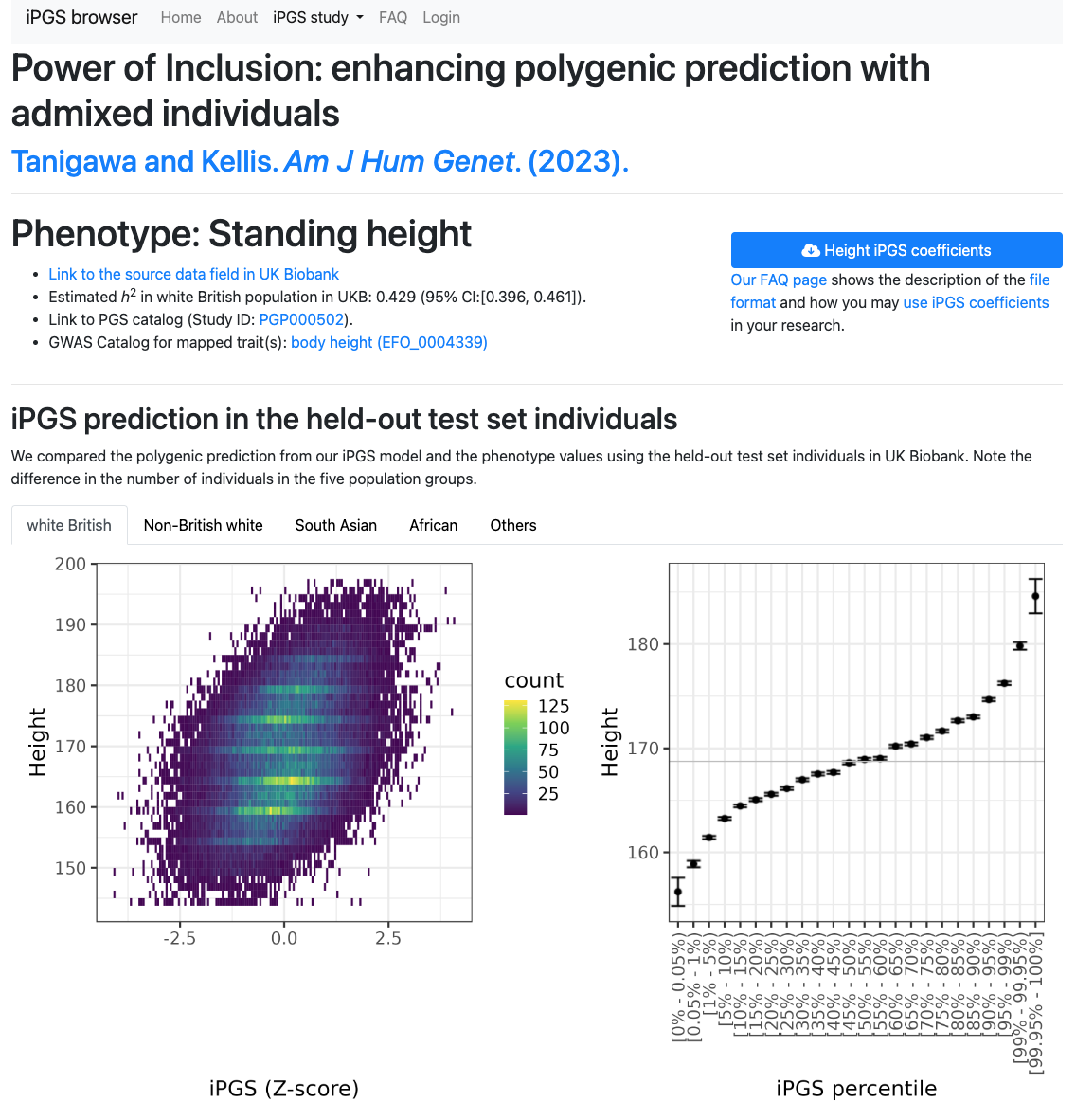

The dataset is also available in the PGS catalog (Study ID: PGP000502) and figshare.
Links
- Tanigawa and Kellis. Am J Hum Genet (2023)
- MIT News article by Anne Trafton
- Inside AJHG: A Chat with Yosuke Tanigawa
- Inclusive Polygenic Scores (iPGS) data browser
- Data @ figshare
- Twitter/X summary:
日本語での解説 (Highlights in Japanese)
近年、ポリジェニック・スコア(PGS)と呼ばれる統計解析技術と、その臨床応用への関心が高まっている。ポリジェニック・スコアは、各個人のゲノムに含まれる数千〜数百万もの遺伝子変異の効果を足し合わせ、個人レベルの疾患リスクや疾患関連形質を予測を行う。アメリカ心臓協会が科学的声明を発表するなどPGSの臨床応用への期待も高まる一方、現在のモデルはヨーロッパ系以外の祖先集団において性能が大幅に低下することが知られていた。
本論文では、複数の祖先集団にルーツをもつミックスの人々にも適用可能なインクルーシブ・ポリジェニック・スコア(iPGS)という手法を開発し、提案手法による全ての祖先集団での性能向上を実証した。これまでの手法は、解析に用いるサンプルをヨーロッパ系・アジア系・アフリカ系などあらかじめ決められた祖先集団に分類することが必須となっており、ミックスの人々は解析対象から除外されることが多かった。一方、私たちは、遺伝的な祖先集団は連続的な量で表されるものであり、ミックスの人々の遺伝情報こそがヨーロッパ集団と非ヨーロッパ集団での予測性能の差を埋める鍵となると考え、ミックスの人々を包摂してポリジェニック・スコアを学習する統計解析手法を確立した。
英国 UK Biobank の約406,000人・60形質のデータへ提案手法を適用したところ、従来手法と比較して大幅な性能向上が見られた。このコホートにはアフリカ系祖先集団は約1.5%しか含まれないが、この集団に対する好中球数の予測精度は、従来手法と比べて100倍に向上し、ヨーロッパ集団に対するものと同等以上の予測性能を達成した。60形質の平均では、アフリカ系集団に対して平均60.8%、南アジア系集団に対して11.6%、ヨーロッパ集団に対して4.8%、ミックスなどその他の祖先を持つ集団に対して17.8%の性能向上を報告した。
提案手法をより祖先的に多様な人類集団から集められた各種の複雑形質のデータセットに適用することで、心臓病・がん・生活習慣病など、多くの疾患の遺伝的リスクが実現できるであろう。予測される疾患リスクが高い集団に対する重点的な予防的介入など成果の社会実装に、我々のよりインクルーシブな提案手法が用いられることで、遺伝的多様性によらず全ての人々がゲノム解析の研究成果を享受できるようになると期待される。
Reference: Tanigawa and Kellis. Power of inclusion: Enhancing polygenic prediction with admixed individuals. The American Journal of Human Genetics. 110, 1888-1902 (2023). https://doi.org/10.1016/j.ajhg.2023.09.013
