Hypometric genetics: Improved power in genetic discovery by incorporating quality control flags
Published in The American Journal of Human Genetics, 2024
We introduce “hypometric genetics,” an approach to investigate the genetic basis of binarized traits representing the presence of below-the-limit-of-quantification (BLQ) quality control indicators. We show a joint analysis of BLQ and quantitative traits leads to improved power in genetic discovery, highlighting the value of considering population-based samples and phenotypic extremes in genetic studies. Links to Article full text and MIT CSAIL News.
Abstract
Balancing the tradeoff between quantity and quality of phenotypic data is critical in omics studies. Measurements below the limit of quantification (BLQ) are often tagged in quality control fields, but these flags are currently underutilized in human genetics studies. Extreme phenotype sampling is advantageous for mapping rare variant effects. We hypothesize that genetic drivers, along with environmental and technical factors, contribute to the presence of BLQ flags. Here, we introduce “hypometric genetics” (hMG) analysis and uncover a genetic basis for BLQ flags, indicating an additional source of genetic signal for genetic discovery, especially from phenotypic extremes. Applying our hMG approach to n = 227,469 UK Biobank individuals with metabolomic profiles, we reveal more than 5% heritability for BLQ flags and report biologically relevant associations, for example, at APOC3, APOA5, and PDE3B loci. For common variants, polygenic scores trained only for BLQ flags predict the corresponding quantitative traits with 91% accuracy, validating the genetic basis. For rare coding variant associations, we find an asymmetric 65.4% higher enrichment of metabolite-lowering associations for BLQ flags, highlighting the impact of putative loss-of-function variants with large effects on phenotypic extremes. Joint analysis of binarized BLQ flags and the corresponding quantitative metabolite measurements improves power in Bayesian rare variant aggregation tests, resulting in an average of 181% more prioritized genes. Our approach is broadly applicable to omics profiling. Overall, our results underscore the benefit of integrating quality control flags and quantitative measurements and highlight the advantage of joint analysis of population-based samples and phenotypic extremes in human genetics studies.
Highlights
Omics profiling coupled with human genetic studies provides excellent opportunities to investigate the genetic basis of disease and intermediate traits. Laboratory measures often include quality control flags, such as below-the-limit-of-quantification (BLQ).
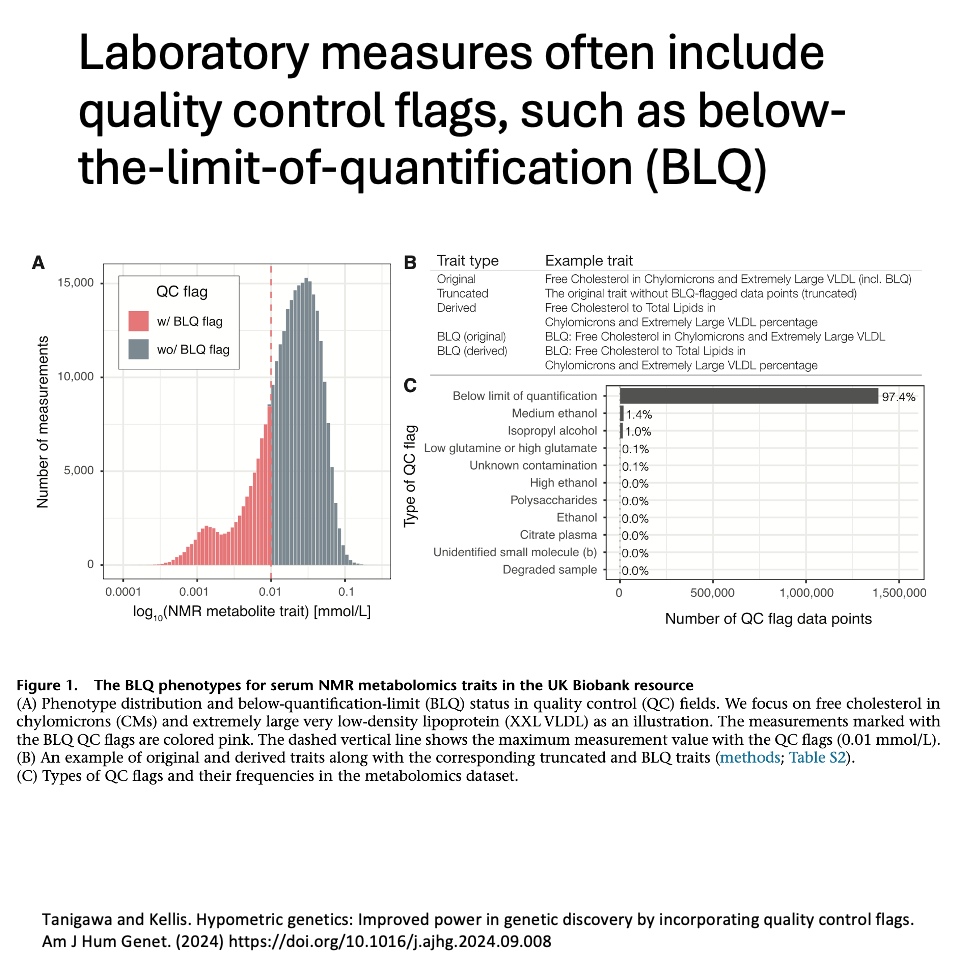
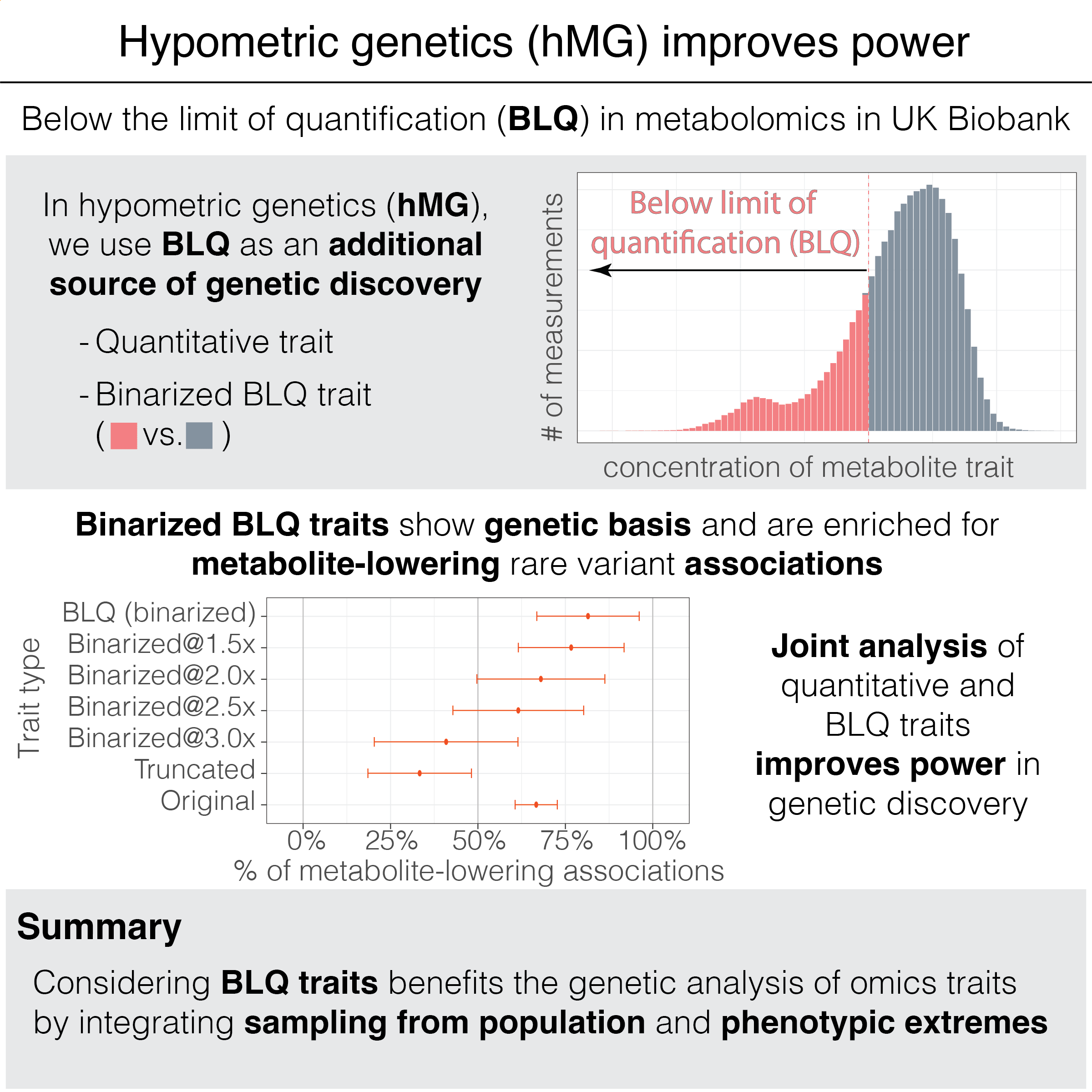
We hypothesize that extremely low measurement values tagged with BLQ flags are influenced by a combination of technical and biological factors. We call those measurements ‘hypometric’ and set out to investigate their genetic basis using metabolomics profiles in UK Biobank.
We revealed that the binarized traits representing the presence or absence of BLQ flags have a genetic basis. One may conduct genetic analyses with the binarized BLQ traits, including common and rare association mapping, polygenic score modeling, and gene prioritization.
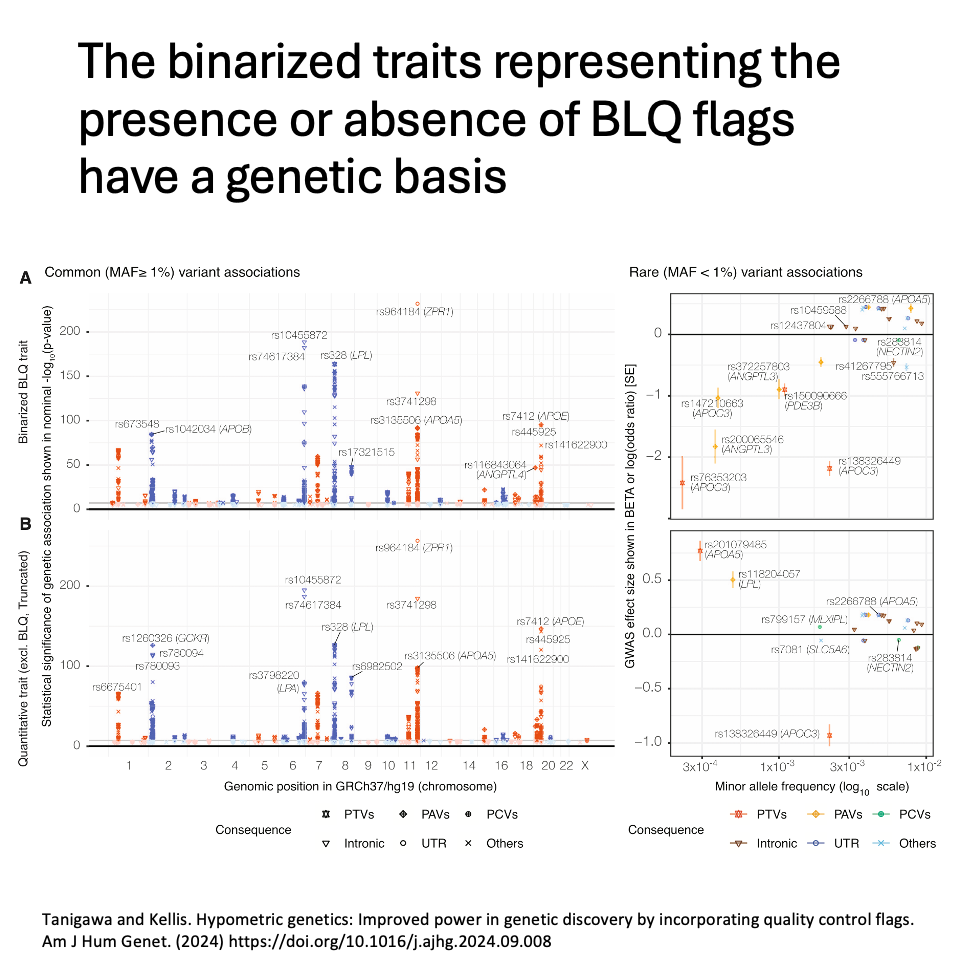
For common variants, genetic associations are similar between the binarized BLQ traits and the corresponding quantitative traits. Polygenic scores trained only on the binarized BLQ traits predict quantitative traits at ~91% accuracy.
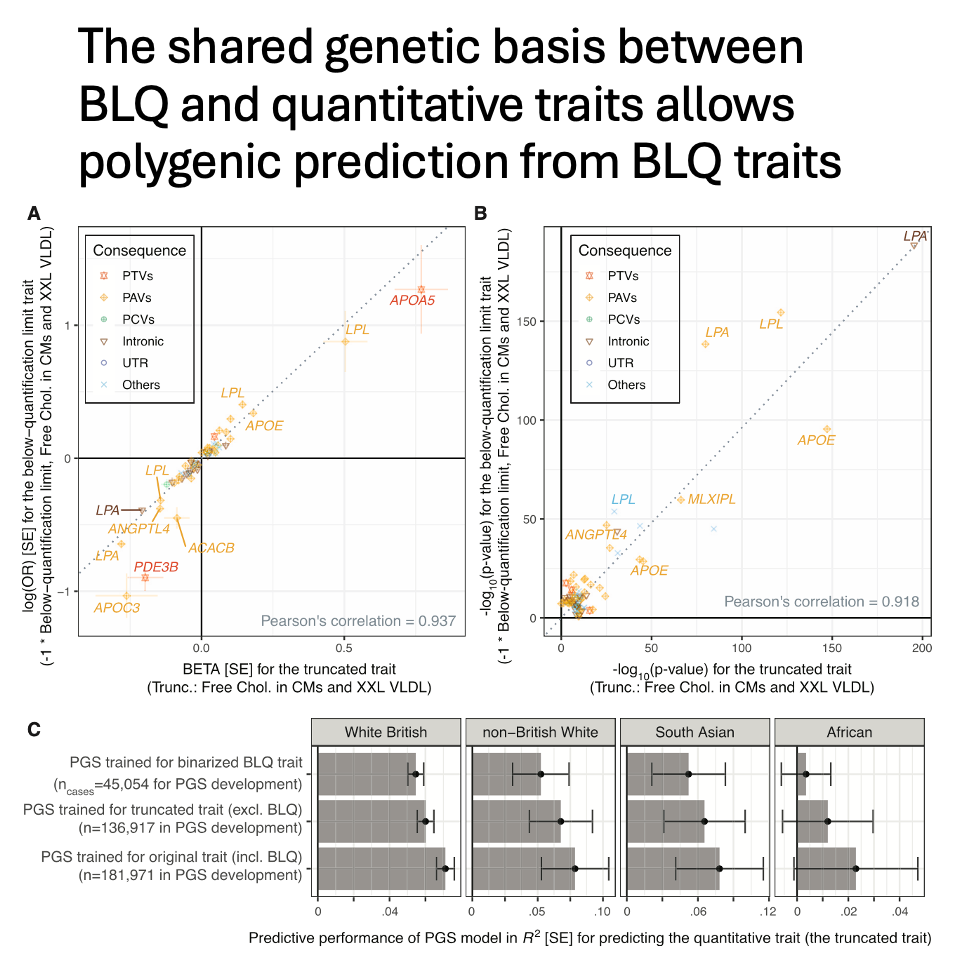
For rare variants, metabolite-lowering effects are enriched for genetic associations in BLQ traits, highlighting the utility of analyzing individuals flagged with BLQ traits: they carry extremely low concentrations of metabolites, i.e., phenotypic extremes.
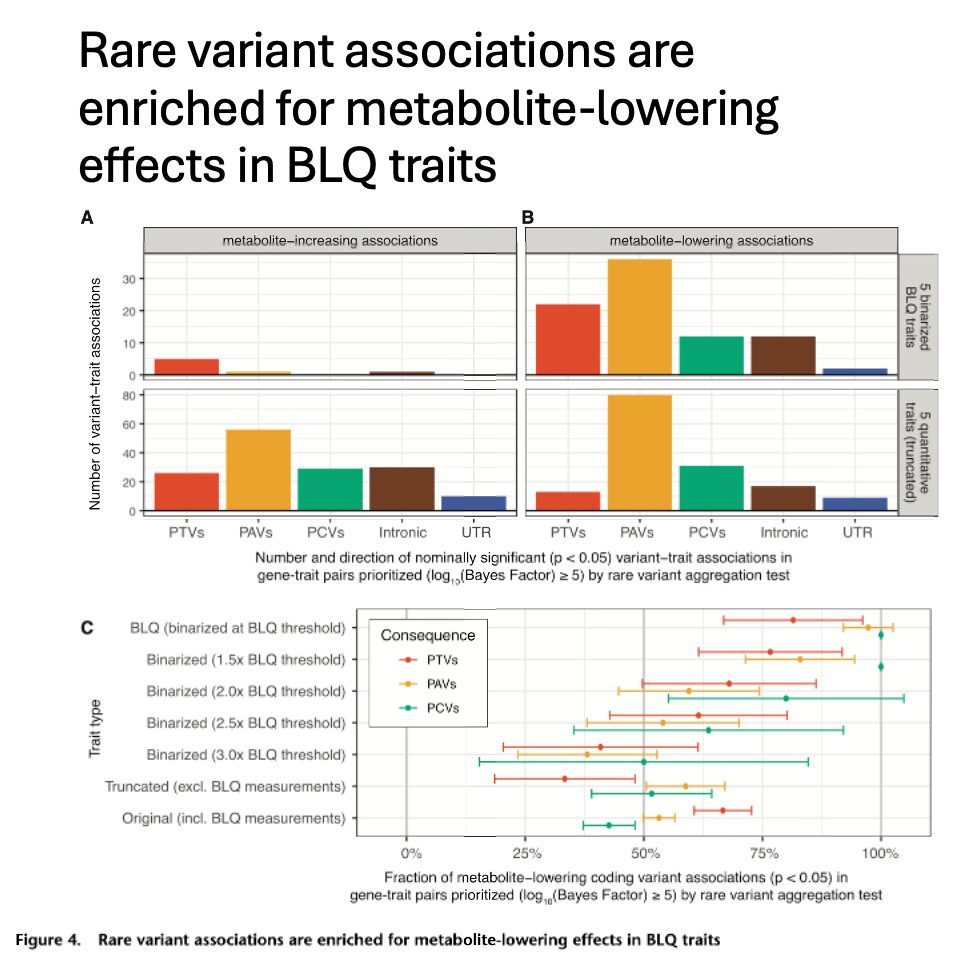
We show that joint analysis of binarized BLQ traits and the corresponding quantitative traits improves the statistical power in genetic discovery, leading to an average of 181% more prioritized genes in Bayesian rare variant aggregation tests.
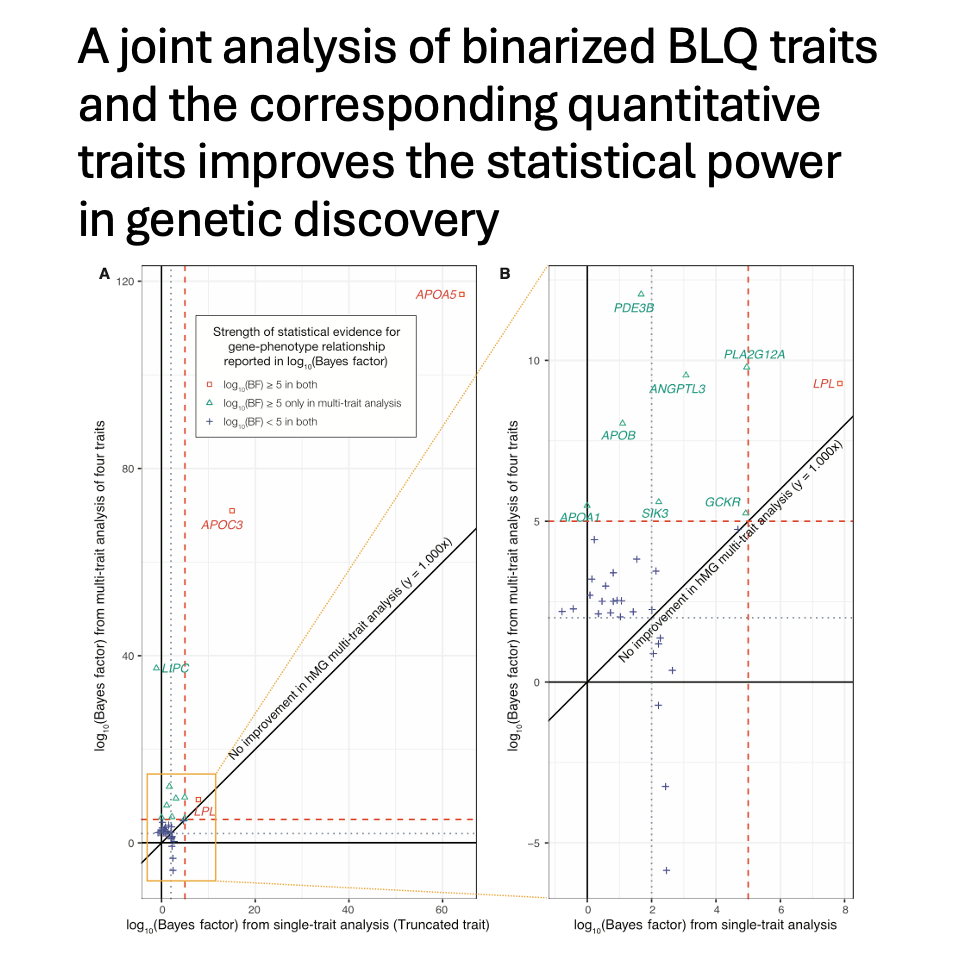
Overall, our results highlight the critical benefits of integrating QC flags with quantitative measurements in human genetics studies. More broadly, our results underscore the benefit of considering the phenotypic extremes and population-based samples in cohort studies. (8/n)
We thank UK Biobank, its participants, amazing colleagues, as well as funding.
The iPGS browser and dataset availability
The dataset is also available in OSFramework. The polygenic score models are also available in the iPGS Browser.
Links
Reference: Tanigawa and Kellis. Hypometric genetics: Improved power in genetic discovery by incorporating quality control flags. The American Journal of Human Genetics. 111(11):2478-2493 (2024). https://doi.org/10.1016/j.ajhg.2024.09.008
